Serological assays for SARS‐CoV‐2 present challenges and opportunities
Timely, scalable and accurate diagnostic testing is crucial in the prevention and control of the coronavirus disease 2019 (COVID‐19) pandemic.1 With limited treatment options and no available vaccine, the accurate and timely identification of infectious patients with COVID‐19 is instrumental to the public health outbreak response. Isolation of patients with COVID‐19, contact tracing and quarantine measures, in addition to physical distancing within the community, have proven effective in reducing case numbers.2 Due to the high sensitivity and specificity in symptomatic individuals, detection of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection by reverse transcriptase polymerase chain reaction (RT‐PCR) is the gold standard method for confirming cases of COVID‐19.3 In contrast, serological assays have lower utility in the initial investigation of suspected cases, but are essential in the development and evaluation of therapeutic agents and to inform modelling and public health policy as this pandemic progresses.
As part of initial laboratory responses, Chinese investigators released the viral whole genome sequence in early January 2020, which enabled the rapid development of RT‐PCR workflows for the detection of SARS‐CoV‐2.4 However, the unprecedented scale of RT‐PCR diagnostic testing has placed extraordinary demands on health care and laboratory systems, with both challenges relating to supply chains of reagents and to the workforce resource required to support population‐level testing. Since the start of the pandemic, a range of commercially available diagnostic tests has been released, including RT‐PCR assays, point‐of‐care and laboratory‐based serological tests. These tests vary both in analytical performance and in their particular utility in the overall public health response to COVID‐19.
Performance aspects of serological tests
Following SARS‐CoV‐2 infection, specific antibodies to different components of this virus are generated. Depending on the antigen target used by the assay, detection of these antibodies (IgM, IgA, IgG or total antibody) may indicate exposure (non‐neutralising antibodies) or potential immunity (neutralising antibodies). To date, a range of serological tests for COVID‐19 have been developed, each with particular test characteristics (Box 1). Broadly, these serological tests can be divided into tests that (i) can be performed at the point‐of‐care; (ii) can be performed in routine diagnostic laboratories, and (iii) can only be performed in specialised reference laboratories (Box 1).
The majority of point‐of‐care and laboratory‐based assays have incorporated either the nucleocapsid antigen (N) or part of the spike protein (S), often the S1 region or the receptor binding domain (RBD). The RBD has been shown to correlate well with the production of neutralising antibodies,5 while some studies have shown N to be immunodominant, producing an earlier or stronger immune response.6 Most patients with COVID‐19 seroconvert by day 10–14 (~ 80%) following the onset of symptoms, with almost 100% seroconversion by day 20.7 However, comparisons across published studies are challenging due to the different antigens used in assays, differences in the complexity of patient populations, variations in the RT‐PCR assays used as the gold standard for determining the sensitivity of serological assays, and often limited data on the timing of sample collection post‐COVID‐19 symptom onset. Further, it is not clear whether the type and amount of antibody correlate with severity of disease or, more importantly, with immune protection from re‐infection. As noted by the World Health Organization, the Australian Public Health Laboratory Network (PHLN) and the Royal College of Pathologists of Australasia (RCPA), a negative result using a serological test does not rule out SARS‐CoV‐2 infection, particularly in individuals with strong epidemiological risk factors, and both the PHLN and the RCPA note that there is no role for serological point‐of‐care tests (PoCT) in the acute diagnosis of COVID‐19.8,9
Point‐of‐care testing
As some of the first COVID‐19 serological assays available, significant publicity accompanied the release of PoCT. PoCT involve detection of anti‐SARS‐CoV‐2 antibodies through binding to immobilised antigens, generally bound to colloidal gold on a test strip (Box 2). The relatively cheap and simple nature of lateral flow assays means that production is suited to scale‐up for increased testing capacity. Post‐market validation studies have demonstrated variable performance characteristics, often inferior to that reported by manufacturers, with sensitivities for IgG reported in the range of 53–100% for samples collected more than 14 days after symptom onset, and specificities of 91.7–100%.10,11 Careful test selection and consideration of the clinical utility before application are therefore critical. The National Pathology Accreditation Advisory Council has existing guidelines on the use of PoCT in Australia.12 These guidelines cover issues such as clinical supervision for performing PoCT, ensuring test quality, staff training and competency for performing PoCT, and appropriate reporting of test results. More recently, this advice has been extended to serological PoCT for COVID‐19, with an emphasis on a robust quality framework to support the implementation and deployment of such tests. Of note, in Australia, the supply of self‐testing kits (eg, testing at home) for many infectious diseases, including COVID‐19, is prohibited under another Therapeutic Goods Administration regulation, the Therapeutic Goods (Excluded Purposes) Specification 2010.13
Laboratory‐based assays
A wide variety of laboratory‐based serological assays are now available, most commonly enzyme immunosorbent assays (ELISA) or chemiluminescent immunoassay (CLIA/CMIA) format. Assays may be semi‐automatic or completely automated, lending themselves to large scale testing and reporting. In general, performance characteristics have been more consistent and closer to that reported by manufacturer's compared with PoCT, with IgG sensitivities in the range of 80–100% for samples collected more than 14 days after symptom onset, and specificity commonly falling between 95% and 100%.10,11,14
Use of serological assays
Given the time lag from symptom onset to detectable antibody, serological PoCT have no role in the detection of acute COVID‐19. However, there are some settings where serological assays, including PoCT, may have potential utility, including defining antibody prevalence in key populations such as frontline workers and determining the extent of COVID‐19 transmission within the community. For other applications, such as identifying individuals for further evaluation of therapeutic immunoglobulin donation and vaccine development and evaluation, assays that assess neutralising antibody response are likely to be required.
Regardless of the type of serological assay used, in order to appropriately deploy serological testing, it is critical to understand the limitations of test performance in the epidemiological context in which tests are used. This is particularly important in a setting such as Australia, where, based on the number of reported cases of COVID‐19 (24 236 cases as of 20 August 2020), there is an estimated COVID‐19 period prevalence of 0.095% (January to August 2020). As such, even with serological tests that are highly sensitive and specific, the majority of positive tests are likely to represent false positive results. When considering the use of serology to inform policies relating to relaxing of physical distancing interventions, the specificity of the assay becomes critical. If most individuals considered immune actually represent false positive results, then the threshold to maintain immunity (if this indeed correlates with antibody detection) within the community will not be achieved. Consideration should therefore be given for confirmation of initial positive results by either retesting on an assay with an alternative target, or retesting with serological gold standard assays, such as microneutralisation or western blot assays.15
Application of serological assays and future research needs
Understanding local transmission dynamics and/or exposure risk through serological surveys can inform local health policy at an institutional, state or national level. For example, a recent large serological survey in Spain, including more than 50 000 residents, used both PoCT and a laboratory‐based CLIA to estimate a seroprevalence across the country of 5.0%, following an initial COVID‐19 outbreak in February to April.16 It was estimated that approximately a third of cases were asymptomatic, while health care workers had a higher seropositivity than the community (10.2% v 5.0%). This is in contrast to health care workers in Belgium, where direct contact with patients with COVID‐19 did not increase the odds of being seropositive.17
The degree and duration of immunity following SARS‐CoV‐2 infection is unknown, but if in keeping with other coronaviruses (approximately 40 weeks), immunity is unlikely to be lifelong and may be shorter lived in milder infections.7 Duration of immunity is a critical area for future research, as it is the key component in models estimating the frequency of SARS‐CoV‐2 infection incidence in the coming years (eg, second or third waves, or annual seasonal COVID‐19 activity similar to influenza), and to determine the utility of policies such as “immunity passports”.18
Conclusion
The unprecedented demands on laboratories to rapidly upscale testing for COVID‐19 has necessarily led to fast‐tracking of normally stringent regulatory requirements for test approval, both globally and in Australia. Following the recent publication of peer‐reviewed high quality validation data, serological testing is now available in many Australian laboratories. Serological testing will complement the current clinical utility of RT‐PCR for SARS‐CoV‐2 infection diagnosis, highlight local transmission dynamics, and further our understanding of what the future brings for the COVID‐19 pandemic.
Box 1 – Main serological assays used to date for the detection of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)
|
Serological assay |
Detection method |
Advantages |
Disadvantages |
Implications |
|||||||||||
|
|
|||||||||||||||
|
Neutralisation |
|
|
|
|
|||||||||||
|
Indirect fluorescent antibody (IFA) |
|
|
|
|
|||||||||||
|
Enzyme immunoassay (EIA) |
|
|
|
|
|||||||||||
|
Lateral flow EIA |
|
|
|
|
|||||||||||
|
|
|||||||||||||||
|
PC2 = physical containment level 2; PC3 = physical containment level 3. |
|||||||||||||||
Box 2 – Schematic of a lateral flow immunoassay for detection of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) IgM and IgG antibodies*
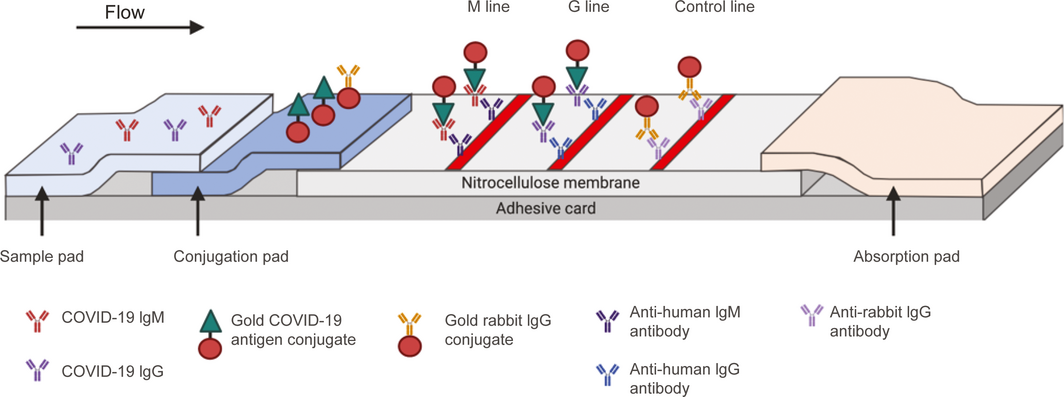
*The sample is added to the sample pad, and then travels by capillary motion to the conjugation pad. Anti‐SARS‐CoV‐2 IgM and/or IgG antibodies in the patient sample then bind to the specific SARS‐CoV‐2 antigen. This antigen is bound to colloidal gold, which acts as a colorimetric indicator. The bound antigen‐antibody‐gold complex then travels to the nitrocellulose membrane and bind to specific anti‐human IgM or IgG antibodies, with a resultant colorimetric change. To monitor test validity, excess conjugated colloidal gold binds to antibody on the control line, which allows assessment of whether the fluid has successfully migrated across the test strip. Source: Adapted from Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM–IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J Med Virol 2020; https://doi.org/10.1002/jmv.25727. [Epub ahead of print]
Provenance: Not commissioned; externally peer reviewed.
- 1. Sharfstein JM, Becker SJ, Mello MM. diagnostic testing for the novel coronavirus. JAMA 2020; 323: 1437.
- 2. Okell LC, Verity R, Watson OJ, et al. Have deaths from COVID‐19 in Europe plateaued due to herd immunity? Lancet 2020; 395: e110–111.
- 3. Public Health Laboratory Network of Australia. PHLN guidance on laboratory testing for SARS‐CoV‐2 (the virus that causes COVID‐19) [website]. PHLN, 2020. https://www.health.gov.au/resources/publications/phln-guidance-on-laboratory-testing-for-sars-coV-2-the-virus-that-causes-covid-19 (viewed July 2020).
- 4. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill 2020; 25: 2000045.
- 5. Quinlan BD, Mou H, Zhang L, et al. The SARS‐CoV‐2 receptor‐binding domain elicits a potent neutralizing response without antibody‐dependent enhancement [preprint]. bioRxiv 2020; https://doi.org/10.1101/2020.04.10.036418.
- 6. Hachim A, Kavian N, Cohen C, et al. Beyond the spike: identification of viral targets of the antibody response to SARS‐CoV‐2 in COVID‐19 patients [preprint]. medRxiv 2020; https://doi.org/10.1101/2020.04.30.20085670.
- 7. Kellam P, Barclay W. The dynamics of humoral immune responses following SARS‐CoV‐2 infection and the potential for reinfection. J Gen Virol 2020; https://doi.org/10.1099/jgv.0.001439. [Epub ahead of print].
- 8. Public Health Laboratory Network Australia. PHLN statement on point‐of‐care serology testing for SARS‐CoV‐2 (the virus that causes COVID‐19) [website]. https://www.health.gov.au/resources/publications/phln-statement-on-point-of-care-serology-testing-for-sars-coV-2-the-virus-that-causes-covid-19 (viewed July 2020).
- 9. Royal College of Pathologists Australasia. Position statement: COVID‐19 IgG/IgM rapid PoCT tests. Sydney: RCPA, 2020. https://www.rcpa.edu.au/getattachment/bf9c7996-6467-44e6-81f2-e2e0cd71a4c7/COVID19-IgG-IgM-RAPID-POCT-TESTS.aspx (viewed July 2020).
- 10. Lisboa Bastos M, Tavaziva G, Abidi SK, et al. Diagnostic accuracy of serological tests for COVID‐19: systematic review and meta‐analysis. BMJ 2020; 370: m2516.
- 11. Deeks JJ, Dinnes J, Takwoingi Y, et al. Antibody tests for identification of current and past infection with SARS‐CoV‐2. Cochrane Database of Syst Rev 2020; (6): CD013652.
- 12. National Pathology Accreditation Advisory Council. Guidelines for point of care testing, 1st ed. Commonwealth of Australia, 2015. https://www1.health.gov.au/internet/main/publishing.nsf/Content/35DE5FC4786CBB33CA257EEB007C7BF2/$File/Guidelines%20PoCT%201st%20Ed%202015.pdf (viewed July 2020).
- 13. Therapeutic Goods Administration. Warning to consumers and advertisers about COVID‐19 test kits [website]. https://www.tga.gov.au/media-release/warning-consumers-and-advertisers-about-covid-19-test-kits (viewed July 2020).
- 14. Public Health England. COVID‐19: laboratory evaluations of serological assays [website]. Public Health England, 2020. https://www.gov.uk/government/publications/covid-19-laboratory-evaluations-of-serological-assays (viewed July 2020).
- 15. Public Health Laboratory Network. Public Health Laboratory Network guidance for serological testing in COVID‐19. Canberra: Department of Health, 2020. https://www1.health.gov.au/internet/main/publishing.nsf/Content/Publications-13 (viewed July 2020).
- 16. Pollán M, Pérez‐Gómez B, Pastor‐Barriuso R, et al. Prevalence of SARS‐CoV‐2 in Spain (ENE‐COVID): a nationwide, population‐based seroepidemiological study. Lancet 2020; https://doi.org/10.1016/S0140-6736(20)31483-5. [Epub ahead of print]
- 17. Steensels D, Oris E, Coninx L, et al. Hospital‐wide SARS‐CoV‐2 antibody screening in 3056 staff in a tertiary center in Belgium. JAMA 2020; 324: 195.
- 18. Kissler SM, Tedijanto C, Goldstein E, et al. Projecting the transmission dynamics of SARS‐CoV‐2 through the postpandemic period. Science 2020; 368: 860–868.





Katherine Bond is supported by a National Health and Medical Research Council (NHMRC) of Australia Postgraduate Scholarship (GNT1191321). Benjamin Howden is supported by an NHMRC Practitioner Fellowship (APP1105905). Deborah Williamson is supported by an NHMRC Investigator Grant (APP1174555).
No relevant disclosures.