Australia has been seriously planning for a pandemic since 2005, when there was concern about the pandemic potential of the H5N1 strain of avian influenza (a virus strain that turned out to have enhanced virulence but was not readily transmissible to humans), with the development of various federal1,2 and state3-10 pandemic plans. With the exception of Queensland (which signed a memorandum of understanding with private pathology laboratories and funded them for testing infrastructure), all jurisdictions decided that a central public reference laboratory testing protocol would be the best response in a pandemic. All guidelines specified that the presence or absence of influenza A infection should be notified to the managing clinician within 24 hours of receipt of a sample to assist in clinical management.1-10 A further 24 hours was specified as the benchmark for determining whether an influenza A specimen represented the pandemic strain. Polymerase chain reaction (PCR) testing was favoured over antigen testing (which is offered by many private sector and peripheral public sector laboratories) because of an assumed superior sensitivity of the former test in an outbreak situation. The public sector Communicable Diseases Network Australia also promulgated similar guidelines.11
The Australian Government commissioned a study by the Health Infrastructure Assurance Advisory Group on private pathology laboratory surge capacity, to determine whether the private sector could provide support during national disasters such as pandemics. The report on the study found that the private sector had considerable underused capacity for molecular diagnosis of influenza and recommended that its resources be included in pandemic plans.12 The ability of the private sector to do this is currently hampered by the Medicare Benefits Schedule’s policy of only reimbursing (at close to cost price) testing for a maximum of three nucleic acids (item 69496). This is not economically viable for most private laboratories, especially if testing for the usual panel of seven to eight viruses is requested (influenza A, influenza B, parainfluenza 1–3, adenovirus, respiratory syncytial virus, and human metapneumovirus). Many guidelines also advise simultaneously requesting PCR assays for atypical agents such as Mycoplasma pneumoniae, Chlamydophila pneumoniae, Chlamydophila psittaci, Legionella pneumophila, Legionella longbeachae, and Coxiella burnetii in some circumstances, as clinical manifestations are rarely characteristic of a particular aetiology. This makes pathology testing even less commercially viable. An expanded panel is nevertheless important because, in managing patients with possible pandemic influenza, identifying an alternative diagnosis is just as helpful in clinical management as determining the pandemic influenza strain status (enabling specific therapy and making patient triage and isolation more efficient).
In December 2007, the public sector Public Health Laboratory Network had a face-to-face meeting with private pathology laboratories to discuss these issues. But, despite these discussions, none of the jurisdictions with plans for centralised testing changed their plans, and pandemic funds were channelled solely to public sector reference laboratories.4-12
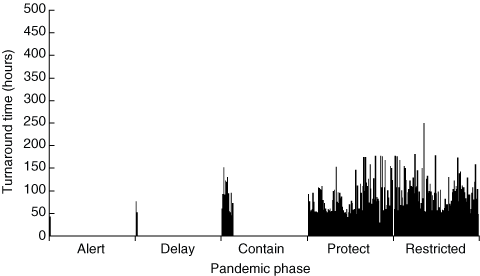
The experience of our laboratory (a private pathology service in Western Australia and the Northern Territory) during the current pandemic may be helpful in determining whether such centralised strategies have been successful thus far. To date, since the declaration by the Australian Government Department of Health and Ageing of the “Alert” phase on 25 April 2009, we have referred over 5500 specimens from patients with suspected influenza to a reference laboratory for diagnostic testing. Of these, 220 have been reported back as positive for influenza A (191 for the pandemic H1N1 strain, three for seasonal H1 influenza A, and 26 for H3 influenza A). Turnaround times for results, calculated from the time of specimen collection to the time of electronic download of the result into our computer system, are charted in Box 1. (These are best-case scenario estimates, as the results also need to be delivered to the requesting doctor by phone, hard copy or electronic transfer.) Although the number of positive results was very low in the initial two phases of the pandemic, it is clear that the turnaround time rose progressively with each phase and that the target turnaround time of 48 hours was only ever achieved in the first phase. Acknowledgement that the reference laboratory (PathWest Laboratory Medicine WA) was not coping with the demand was confirmed by communiqués released on 12 June 2009 (discouraging testing for uncomplicated cases) and 22 July 2009 (notifying doctors that specimens from high-risk cases only would be processed). This later period is represented by the last column in Box 1 (the “restricted” phase). In spite of these measures, there has not yet been any improvement in test turnaround time. During this period, the thermocycler (PCR machine) in our laboratory (and presumably in many other private pathology laboratories) was relatively underused. It would appear that similar experiences were noted in other jurisdictions,13 but it would be interesting to know whether the Queensland experience was modified by their different approach.
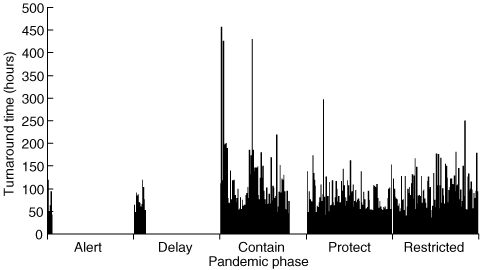
For illustrative purposes only, total turnaround times for a random sample of 409 influenza A-positive and -negative specimens are displayed in Box 2. These data showed a similar pattern to the positive-only specimens, with rising turnaround times as the pandemic phases progressed, except that the benchmark was not achieved even during the first phase. This is presumably because patients at the highest risk of disease (who were the most likely to test positive for influenza A) had specimen transport facilitated by use of a taxi as well as better coordination with the recipient laboratory’s testing runs.
[T]here is no clear articulation [in the document] of the fundamental need for close interaction between public and private sectors to address the threat posed by a pandemic.
The Australian Government was also advised at the time that
Given that most health interactions in Australia occur in the community (where community general practitioners consult and send appropriate laboratory requests to private pathology laboratories), the initial cases in a pandemic will almost certainly present there. In addition, very shortly after the establishment of a pandemic in Australia, the public hospital system and their diagnostic laboratories will effectively be overwhelmed and potentially grind to a halt.
Ah, “The best laid schemes o’ mice and men gang aft agley!” (Robert Burns).
- Miles H Beaman1
- Michael J Leung2
- Department of Microbiology, Western Diagnostic Pathology, Perth, WA.
We would like to thank Dr Desmond Chih and Ms Sylvia Gandossi for assistance with data collection and graphics.
We are both employees of Symbion Health, a private sector pathology company.
- 1. Australian Department of Health and Ageing. Interim national pandemic influenza clinical guidelines (June 2006). Canberra: Commonwealth of Australia, 2006. http://www.flupandemic.gov.au/internet/panflu/publishing.nsf/Content/9D4CC1F412DCC346CA2573D70001B875/$File/pandemic-clinical-gl.pdf (accessed Aug 2009).
- 2. Australian Department of Health and Ageing. Australian health management plan for pandemic influenza 2008. Canberra: Commonwealth of Australia, 2008. http://www.health.gov.au/internet/panflu/publishing.nsf/Content/8435EDE93CB6FCB8CA2573D700128ACA/$File/Pandemic%20FINAL%20webready.pdf (accessed Aug 2009).
- 3. Queensland Government. Queensland pandemic influenza plan 2009. Brisbane: Queensland Government, 2009. http://www.premiers.qld.gov.au/publications/categories/plans/assets/pandemic-influenza-plan-2009.pdf (accessed Aug 2009, link updated 8 January 2010).
- 4. Department of Health South Australia. Pandemic influenza: a summary of Health’s operational plan. Adelaide: Government of South Australia, 2007. http://www.pandemicinfluenza.sa.gov.au/LinkClick.aspx?fileticket=H6wF0tzp9x8%3d&tabid=61 (accessed Aug 2009).
- 5. Department of Health, Government of Western Australia. Western Australian health management plan for pandemic influenza 2009. Perth: Government of Western Australia, 2009. http://www.public.health.wa.gov.au/cproot/2233/2/Western%20Australian%20Health% 20Management%20Plan%20for%20Pandemic%20Influenza%202009.pdf (accessed Aug 2009).
- 6. Victorian Department of Human Services. Victorian health management plan for pandemic influenza. Melbourne: State of Victoria, 2007. http://www.health.vic.gov.au/__data/assets/pdf_file/0017/54503/Victorian_health_management_plan_for_pandemic_influenza.pdf (accessed Aug 2009).
- 7. NSW Health. Pandemic preparedness. http://www.health.nsw.gov.au/publichealth/pandemic/index.asp (accessed Aug 2009).
- 8. ACT Health. Australian Capital Territory health management plan for pandemic influenza 2007. Canberra: ACT Health, 2007. http://www.health.act.gov.au/c/health?a=dlpol&policy=1197436800&did=10107160 (accessed Aug 2009).
- 9. Department of Health and Families, Northern Territory Government. Special counter disaster plan. Human pandemic influenza. Darwin: NT Government, 2009. http://www.health.nt.gov.au/library/scripts/objectifyMedia.aspx?file=pdf/31/59.pdf&siteID=1&str_title=NT%20Special %20Counter%20Disaster%20Plan%20for%20Human%20Pandemic%20Influenza.pdf (accessed Aug 2009).
- 10. Government of Tasmania. Tasmanian pandemic (H1N1) 2009 information. http://www.pandemic.tas.gov.au (accessed Aug 2009).
- 11. Communicable Diseases Network Australia. Draft version. Pandemic influenza (PI) response protocol for public health units. http://www.public.health.wa.gov.au/cproot/2250/2/CDNA%20Public%20health%20 response%20protocol.pdf (accessed Aug 2009).
- 12. Australian Department of Health and Ageing. Health emergency preparedness and response. Critical Infrastructure Protection Health Infrastructure Assurance Advisory Group (HIAGG). http://www.health.gov.au/internet/main/publishing.nsf/Content/hepr-hiagg.htm (accessed Aug 2009).
- 13. Grayson ML, Johnson PDR. Australia’s influenza containment plan and the swine flu epidemic in Victoria. Med J Aust 2009: 191: 150. <MJA full text>





Abstract
Australian federal and state governments were advised several years ago that an influenza pandemic would overwhelm Australian public reference laboratories.
It was proposed at the time that currently underused capacity in the private sector be used to enhance pandemic responses.
The current outbreak of pandemic influenza has confirmed the predictions of advisors from the private sector.
Future official pandemic plans should be adjusted to take into account these observations.