Big clinical breakthroughs begin with basic science
The 2003 Nobel Prize in Medicine or Physiology was awarded to a chemist, Paul Lauterbur (US), and a physicist, Peter Mansfield (UK), for their discoveries concerning magnetic resonance imaging,1 while the Nobel Prize in Chemistry was awarded to two medical graduates: Peter Agre (US) for the discovery of water channels and Roderick MacKinnon (US) for structural and mechanistic studies of ion channels.1 The prizes were awarded for seminal discoveries that have had major impacts on medicine and biology. The fascinating story of these discoveries illustrates the value of pursuing basic science research and how breakthroughs at a fundamental level can lead to revolutionary advances in medical care.
In the early 1970s, Paul Lauterbur (at the State University of New York at Stony Brook, US) surmised that if one could vary the magnetic field across a sample in a defined way then it should be possible, by measuring the frequency at which a given ensemble of nuclei were precessing, to determine the exact location of the ensemble in the sample. Lauterbur’s first images were of a “phantom sample” of two tubes of water located within a larger tube of heavy water (ie, water in which the hydrogen atoms are replaced with deuterium atoms).2 Heavy water is chemically almost identical to water and to the eye is indistinguishable. But deuterium nuclei are very different from hydrogen nuclei, so they give no signal when radio waves at the frequency absorbed by hydrogen are applied.
By applying a graded magnetic field across the entire sample, Lauterbur reconstructed an accurate cross-sectional image of the two water-filled tubes (Box 1a). At the time this seemed a rather esoteric experiment, but it was a wonderful model to use for the human body, which is essentially a series of water-filled tubes (eg, arteries) and containers (eg, organs) located within a larger water-filled tube (ie, the body walls).
Lauterbur’s water-filled tubes were geometrically simple, making it relatively easy to solve the mathematics required to reconstruct the images from the radiofrequency output. Although Peter Mansfield (at the University of Nottingham, UK) was working on a different physical system, it was he who was responsible for sorting out the complicated mathematics required to derive images rapidly from NMR spectrometers, thereby making magnetic resonance imaging a feasible clinical application. He also developed the technique of very fast gradient variations (so called echo-planar scanning) that enabled very rapid imaging.3
The journey from the first images of tubes of water to the installation of the first magnetic resonance imaging (MRI) scanner in a hospital required a huge amount of research and development both in the private and public sector, but it only took about 10 years. This involved mathematicians, physicists, engineers, computer scientists, biomedical scientists and medical practitioners in one of the best modern examples of “translational research”. It is estimated that in 2002 about 60 million MRI examinations were performed throughout the world. MRI has had a major impact on many aspects of clinical practice; probably most notably in neurology (Box 1b).
We now know that channels that permit the selective flow of water across cell membranes (aquaporins) are present in almost all cells, but they were only definitively identified 15 years ago. Why did it take so long for them to be discovered? In a sense it was because they are so difficult to measure and because they are so common. Often the easiest way to identify something is to compare two similar objects (in this case, cells) and then determine what it is that is different between them. This of course was not possible for water channels, as almost all cells have them. In the late 1980s, Peter Agre, while working on the rhesus blood group antigens at Johns Hopkins University, US, serendipitously discovered a new membrane protein that he called CHIP28 (channel integral membrane protein of molecular weight 28k). At the time he had no idea what it did.4 Previously and independently, Gheorghe Benga and his group in Romania5 had shown that the water transport inhibitor p-chloromercuribenzoate is selectively bound to a protein in red blood cell membranes. Subsequent studies showed that this was a glycosylated form of CHIP28.
After extensive analysis of the CHIP28 protein Agre’s group recognised that it must form a channel in the red blood cell membrane. The crucial experiment they performed was to over-express the new protein in another cell type (the Xenopus oocyte); they found that the cells became much more permeable to water.6 Since then, Agre and colleagues have shown that there is a whole family of genetically related aquaporins in almost every cell type,7 with some also permeable to glycerol and urea.
In addition to all fluid transporting epithelia, one obvious place where water channels serve a crucial function is in the kidney, where they permit the reabsorption of hundreds of litres of water that pass through the renal glomeruli each day.7
Discovering the channels was one thing, but how do they work? How can these channels allow the passage of water at a very high rate but not allow the passage of hydrated protons (ie, hydronium ions, H3O+)? This question was answered by Agre in collaboration with Fujiyoshi’s group in Kyoto and Engel’s group in Basel when they solved the structure of aquaporin-1 using electron microscopy (Box 2, a,b).8
In contrast to water channels, those that selectively allow the transmembrane flux of ions, such as sodium, potassium or calcium, were first implied in a functional assay and theoretical simulation over 60 years ago, and genes encoding for ion channels were first cloned over 20 years ago.9 We now know that there are hundreds of different ion channels in human cells. Despite extensive analysis of ion channel function, one of the most intriguing conundrums has been how these proteins allow the selective passage of one type of ion at very high rates, while preventing the passage of all others. In the mid 1990s, after electrophysiological measurements combined with molecular biological manipulations of ion channel proteins initially made in Christopher Miller’s laboratory and later in his own laboratory at Harvard University, Roderick MacKinnon realised that the only way to answer this question was to determine the structure of an ion channel using x-ray crystallography. Determining the structure of membrane proteins is extraordinarily difficult, and many people suspected it would not be possible for an ion channel. However, in 1998, MacKinnon and his team (having moved to Rockefeller University) succeeded in crystallising the bacterial potassium channel, KcsA;10 and, just as they had hoped, the structure provided the answer to the riddle of how potassium channels permit the selective passage of potassium ions at a high rate (Box 2, c,d). Furthermore, the combination of x-ray crystallography, electrical measurements and mathematical simulations enabled them to make fundamental predictions about how the channels select ions and open and close to regulate their passage.11 The selectivity of ion channels and the exquisite control of their opening and closing is central to the role they play, for example, in neurotransmission and controlling the heart beat. Indeed, recently it has been shown that even subtle mutations in the genes that encode ion-channel proteins are a potent cause of diseases including epilepsy and cardiac arrhythmias.12
The stories behind the discoveries that lead to the award of a Nobel Prize are always fascinating, while the act of singling out individuals among many who have contributed to a field of research is frequently controversial. However, this should not distract from the real message behind the Nobel Prizes in Medicine or Physiology and Chemistry, which is that pursuing basic science research has enormous practical value. Discoveries made regarding the fundamental properties of molecules, whether by physicists, chemists or biologists, can and do lead to major advances in medical care. Particularly at a time when researchers in Australia are under pressure to pursue applied research at the expense of basic science, this year’s Nobel Prizes provide a potent reminder of the importance of fundamental research.
1: Magnetic resonance imaging
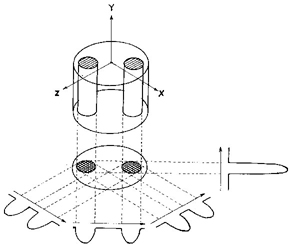  a) Lauterbur’s first published image of a section of a water phantom. Left: model of the phantom. Right: the reconstructed magnetic resonance image (reproduced from reference 2). |
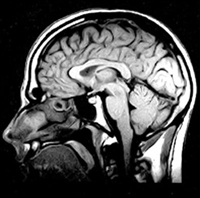 b) Modern magnetic resonance sagittal section of a human head. |
2: Ion channels
a) Ribbon diagram showing structure of aquaporin-1 (water channel). 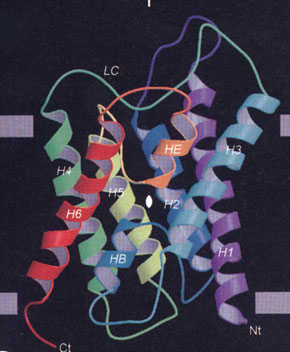 |
b) Model illustrating mechanisms of water permeation in a water channel. The positive charge on the wall of the channel repels hydronium ions, thereby excluding them from crossing the transmembrane region (from reference 8, reproduced with permission of Nature Publishing Group). 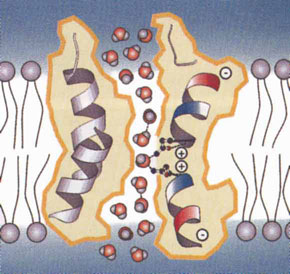 |
c) Structure of the KcsA potassium channel. The channel is a tetramer, but only two subunits are shown, illustrating the cavity and the narrow selectivity filter region formed by the pore helices, labelled ‘P’ (from reference 11, reproduced with permission of Nature Publishing Group). 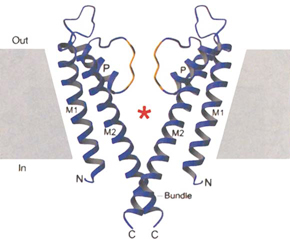 |
d) Model illustrating K+ permeation. The orientation of the pore helices is such as to produce an electronegative well in the cavity, which attracts K+ ions. The narrow selectivity filter region is just the right diameter to accommodate dehydrated K+ ions. K+ permeation occurs in single file. As one K+ ion enters the selectivity filter it repels the K+ ion ahead of it (from reference 10, reproduced with permission of the American Academy for the Advancement of Science). 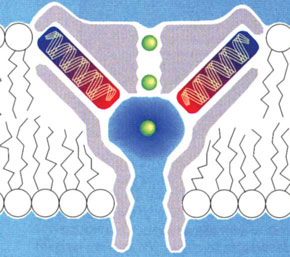 |
Received 16 October 2003, accepted 31 October 2003
- Jamie I Vandenberg1
- Philip W Kuchel2
- 1 Electrophysiology and Biophysics Unit, Victor Chang Cardiac Research Institute, Darlinghurst, NSW.
- 2 Department of Biochemistry, University of Sydney, Sydney, NSW.
- 1. Nobel e-Museum. Official website of the Nobel Foundation. Available at: www.nobel.se (accessed Oct 2003).
- 2. Lauterbur PC. Image formation by induced local interactions: examples employing nuclear magnetic resonance. Nature 1973; 242: 190-191.
- 3. Mansfield P, Maudsley AA. Line scan proton spin imaging in biological structures by NMR. Phys Med Biol 1976; 21: 847-852.
- 4. Denker BM, Smith BL, et al. Identification, purification, and partial characterization of a novel Mr 28,000 integral membrane protein from erythrocytes and renal tubules. J Biol Chem 1988; 263: 15634-15642.
- 5. Benga GH, Popescu O, Pop VI, Holmes RP. Chloromercuribenzenesulfonate binding by membrane proteins and the inhibition of water transport in human erythrocytes. Biochemistry 1986; 25: 1535-1538.
- 6. Preston GM, Carroll TP, Guggino WB, Agre P. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science 1992; 256: 385-387.
- 7. Agre P, King LS, Yasui M, et al. Aquaporin water channels — from atomic structure to clinical medicine. J Physiol 2002; 542: 3-16.
- 8. Murata K, Mitsuoka K, Hirai T, et al. Structural determinants of water permeation through aquaporin-1. Nature 2000; 407: 599-605.
- 9. Noda M, Shimizu S, Tanabe T, et al. Primary structure of Electrophorus electricus sodium channel deduced from cDNA sequence. Nature 1984; 312: 121-127.
- 10. Doyle D, Cabral J, Pfuetzner R, et al. The structure of the potassium channel: molecular basis of K+ conduction and sensitivity. Science 1998; 280: 69-77.
- 11. Jiang Y, Lee A, Chen J, et al.The open pore conformation of potassium channels. Nature 2002; 417: 523-526.
- 12. Ashcroft FM. Ion channels and disease: channelopathies. Academic Press, 2000 (ISBN: 0120653109).




