Coeliac disease is a lifelong, systemic inflammatory disease triggered by dietary gluten.1 Expeditious diagnosis and treatment can reduce morbidity and the impact on quality of life, but many people with the disorder have not been tested and consequently have not been diagnosed.2 A family history of coeliac disease is the strongest risk factor,3 and active case finding among people in at‐risk groups is appropriate. A meta‐analysis found that the pooled prevalence of coeliac disease among 10 252 first degree relatives of people with diagnosed coeliac disease was 7.5%, but the local prevalence differed widely between Asia, Europe, North America, and South America.4
As no relevant Australian data were available, we undertook a prospective observational study at the Wesley Research Institute, Brisbane. First degree relatives of people with diagnosed coeliac disease (most were patients at our clinic) were invited to undergo HLA‐DQ2/8/7 polymerase chain reaction genotyping for coeliac disease risk alleles and were evaluated for serum anti‐tissue transglutaminase (tTG) IgA and anti‐deamidated gliadin peptide (DGP) IgG. When possible, small bowel biopsies were collected from participants with high serum anti‐tTG IgA or anti‐DGP IgG levels to assess anatomical changes. The UnitingCare Health human research ethics committee approved the study (2017.03.217), including access to the participants’ histology results (amendment application #7).
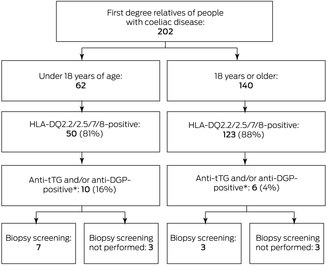
A total of 202 first degree relatives (children, siblings, or parents of 134 people with coeliac disease; no invited relatives declined participation) were screened during 1 June 2017 – 31 March 2019 (81 male, 121 female relatives; median age, 37.3 years; interquartile range, 13.2–47.1 years) (Box 1, Box 2). One family included five first degree relatives (ie, five participants related to one person with coeliac disease), two families included four first degree relatives, eighteen families included three first degree relatives, and 22 families included two first degree relatives; 91 participants were each related to different people with coeliac disease.
Coeliac disease susceptibility haplotypes were detected in 173 participants (86%; 50 children, 123 adults), including 31 children (50%) and 74 adults (53%) with high risk genotypes (Box 3).5 Serology results (anti‐tTG IgA and/or anti‐DGP IgG) were positive for sixteen people with and none without susceptibility haplotypes (Box 4). Small bowel biopsies were collected from seven of the ten children and three of the six adults with positive serology results; diagnostic changes characteristic of coeliac disease (villous atrophy, crypt hyperplasia, intraepithelial lymphocytosis) were determined in all seven children and two of the adults (Box 5),6 each of whom had high risk alleles (DQ2.5). As serological anti‐tTG IgA/anti‐DGP IgG levels for the three children with positive serologic results without biopsies were less than twice the upper limit of normal (ULN), coeliac disease could not be confirmed. In the three adults with positive serologic results who did not undergo biopsy, the anti‐tTG IgA level was at the ULN and the anti‐DGP IgG level normal for one, the anti‐tTG IgA was normal and the anti‐DGP IgG level four times the ULN for the second, and the anti‐tTG IgA was three times the ULN and the anti‐DGP IgG level twice the ULN for the third (Box 4).
In summary, seven of 62 child first degree relatives of people with coeliac disease had biopsy‐confirmed disease, yielding an estimated prevalence of 11%, and a prevalence of 14% for those with coeliac disease susceptibility haplotypes. Two of 140 adult first degree relatives had biopsy‐confirmed disease, yielding an estimated prevalence of coeliac disease of at least 1.4%.
We report the first Australian study to estimate the prevalence of coeliac disease in first degree relatives of people with coeliac disease, as determined by active case finding. The overall seroprevalence of coeliac disease is 1.4%, both globally7 and in Australia.2 The high prevalence we found in children but not adults may reflect delayed diagnosis in Australia.8 That not all participants with positive serology results underwent small bowel biopsy was a limitation of our study, as was the fact that it was a single centre study. Further, we did not include a non‐first degree relatives group, instead comparing our findings with published prevalence reports.
Our findings support active case finding among first degree relatives of people diagnosed with coeliac disease, consistent with overseas guideline recommendations.9,10,11
Box 1 – Testing of first degree relatives of people with coeliac disease for possible coeliac disease
tTG = tissue transglutaminase; DGP = deamidated gliadin peptide.* Threshold for positive serology test result: 7 U/mL.
Box 3 – HLA‐DQ2/8/7 polymerase chain reaction genotyping results for 202 first degree relatives of people with coeliac disease, by age group and risk classification6
|
|
Age group (years) |
||||||||||||||
|
HLA haplotype |
Under 18 |
18 or older |
|||||||||||||
|
|
|||||||||||||||
|
Participants |
62 |
140 |
|||||||||||||
|
No risk expression |
12 (19%) |
17 (12%) |
|||||||||||||
|
High risk (DQ2.5) |
31 (50%) |
74 (53%) |
|||||||||||||
|
2.5/2.5* |
3 |
7 |
|||||||||||||
|
2.5/2.2* |
4 |
4 |
|||||||||||||
|
2.5/8 |
2 |
17 |
|||||||||||||
|
2.5/7 |
5 |
8 |
|||||||||||||
|
2.5/other |
17 |
38 |
|||||||||||||
|
Intermediate risk (DQ8) |
9 (15%) |
18 (13%) |
|||||||||||||
|
2.2/8 |
2 |
3 |
|||||||||||||
|
8/8 |
1 |
1 |
|||||||||||||
|
8/7 |
1 |
0 |
|||||||||||||
|
8/other |
5 |
14 |
|||||||||||||
|
Low risk (DQ2.2/DQ7) |
10 (16%) |
31 (22%) |
|||||||||||||
|
2.2/2.2 |
2 |
4 |
|||||||||||||
|
2.2/7 |
1 |
1 |
|||||||||||||
|
2.2/other |
5 |
15 |
|||||||||||||
|
7/other |
2 |
11 |
|||||||||||||
|
|
|||||||||||||||
|
* Haplotypes conferring highest risk. |
|||||||||||||||
Box 4 – Anti‐tissue transglutaminase (tTG) IgA and anti‐deamidated gliadin peptide (DGP) IgG serology and small bowel biopsy results for 202 first degree relatives of people with coeliac disease who had coeliac disease susceptibility haplotypes
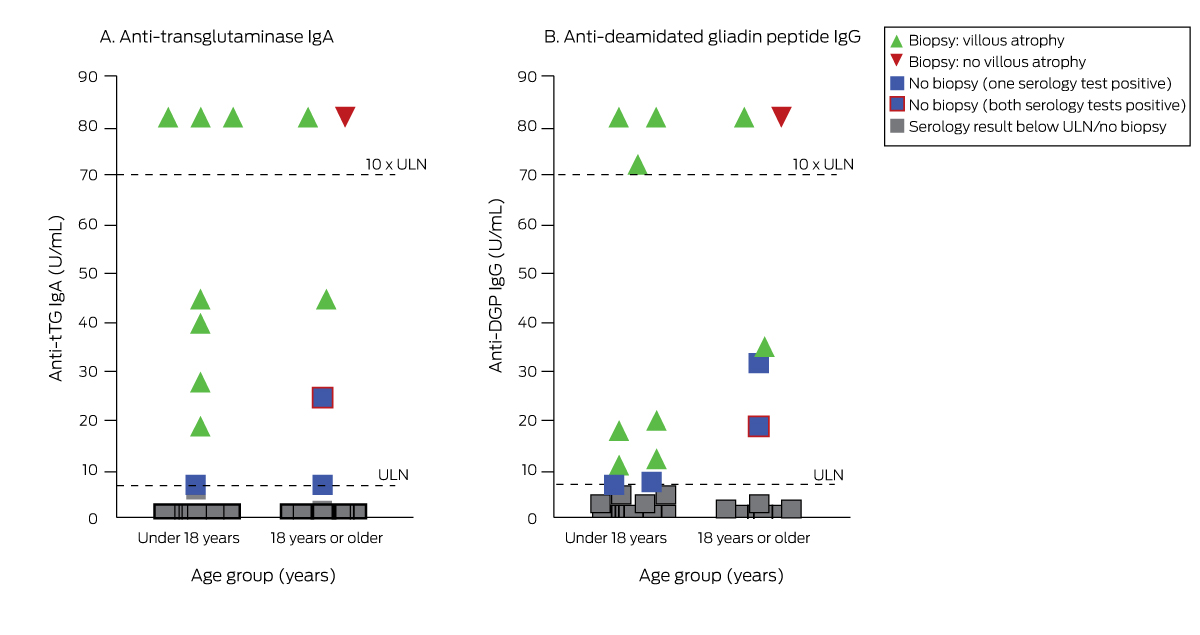
ULN = upper limit of normal (7 U/mL serum).
Box 5 – Modified Marsh–Oberhuber histological classification5 of the ten first degree relatives who underwent duodenal biopsies in our study
|
|
Age group (years) |
||||||||||||||
|
Marsh type |
Under 18 |
18 or older |
|||||||||||||
|
|
|||||||||||||||
|
0 (< 40 IEL/100 enterocytes, normal crypt and villi architecture) |
0 |
1 |
|||||||||||||
|
1 (> 40 IEL/100 enterocytes, normal crypt and villi architecture) |
0 |
0 |
|||||||||||||
|
2 (> 40 IEL/100 enterocytes, crypt hyperplasia, normal villi) |
0 |
0 |
|||||||||||||
|
3a (> 40 IEL/100 enterocytes, crypt hyperplasia, partial villous atrophy) |
2 |
0 |
|||||||||||||
|
3b (> 40 IEL/100 enterocytes, crypt hyperplasia, subtotal villous atrophy) |
3 |
1 |
|||||||||||||
|
3c (> 40 IEL/100 enterocytes, crypt hyperplasia, total villous atrophy) |
2 |
1 |
|||||||||||||
|
|
|||||||||||||||
|
IEL = intraepithelial lymphocytes. |
|||||||||||||||
Received 4 May 2023, accepted 22 August 2023
- 1. Catassi C, Verdu EF, Bai JC, Lionetti E. Coeliac disease. Lancet 2022; 399: 2413‐2426.
- 2. Anderson RP, Henry MJ, Taylor R, et al. A novel serogenetic approach determines the community prevalence of celiac disease and informs improved diagnostic pathways. BMC Med 2013; 11: 188.
- 3. Elwenspoek MMC, Thom H, Sheppard A, et al. Defining the optimum strategy for identifying adults and children with coeliac disease: a systematic review and economic modelling. Health Technol Assess 2022; 26: 1‐310.
- 4. Singh P, Arora S, Lal S, et al. Risk of celiac disease in the first‐ and second‐degree relatives of patients with celiac disease: a systematic review and meta‐analysis. Am J Gastroenterol 2015; 110: 1539‐1548.
- 5. Oberhuber G, Granditsch G, Vogelsang H, et al. The histopathology of celiac disease: time for a standardized report scheme for pathologists. Eur J Gastroenterol Hepatol 1999; 11: 1185‐1194.
- 6. Martínez‐Ojinaga E, Moline M, Polanco I, et al. HLA‐DQ distribution and risk assessment of celiac disease in a Spanish center. Rev Esp Enferm Dis 2018;1107: 421‐426.
- 7. Singh P, Arora A, Strand TA, et al. Global prevalence of celiac disease: systematic review and meta‐analysis. Clin Gastroenterol Hepatol 2018; 16: 823‐836.
- 8. Tye‐Din. Interpreting tests for coeliac disease: tips, pitfalls and updates. Aust J Gen Pract 2018; 47: 28‐33.
- 9. Downey L, Houten R, Murch S, Longson D; Guideline Development Group. Recognition, assessment, and management of coeliac disease: summary of updated NICE guidance. BMJ 2015; 351: h4513.
- 10. Al‐Toma A, Volta U, Auricchio R, et al. European Society for the Study of Coeliac Disease (ESsCD) guideline for coeliac disease and other gluten‐related disorders. United European Gastroenterol J 2019; 7: 583‐613.
- 11. Rubio‐Tapia A, Hill ID, Kelly CP, et al; American College of Gastroenterology. ACG clinical guidelines: diagnosis and management of celiac disease. Am J Gastroenterol 2013; 108: 656‐676.






Open access:
Open access publishing facilitated by The University of Queensland, as part of the Wiley – The University of Queensland agreement via the Council of Australian University Librarians.
This analysis was supported by direct funding for the Coeliac Disease and Immune Health Program at Wesley Research Institute.
No relevant disclosures.