Second infections with the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) are thought to affect fewer than 1% of people with resolved coronavirus disease 2019 (COVID‐19).1 Reinfections as soon as 26 days after the initial diagnosis have been reported, in some cases with increased disease severity.1,2 No confirmed cases of second SARS‐CoV‐2 infections have been reported in Australia, but public awareness of the possibility is needed to encourage continued testing and vaccination.
In late July 2021, three people in one Melbourne household were diagnosed with COVID‐19. There had been a large COVID‐19 outbreak at the workplace of patients 1 and 2, and patient 1 had worked during their acquisition period (ie, the 14 days prior to symptom onset) while infectious co‐workers were present. All three patients had also been diagnosed with COVID‐19 in July 2020, during a period of high community transmission in Victoria; the source of these earlier infections was unknown. Polymerase chain reaction (PCR) cycle threshold (CT) values were consistent with recent infections in both 2020 and 2021 (Box 1).
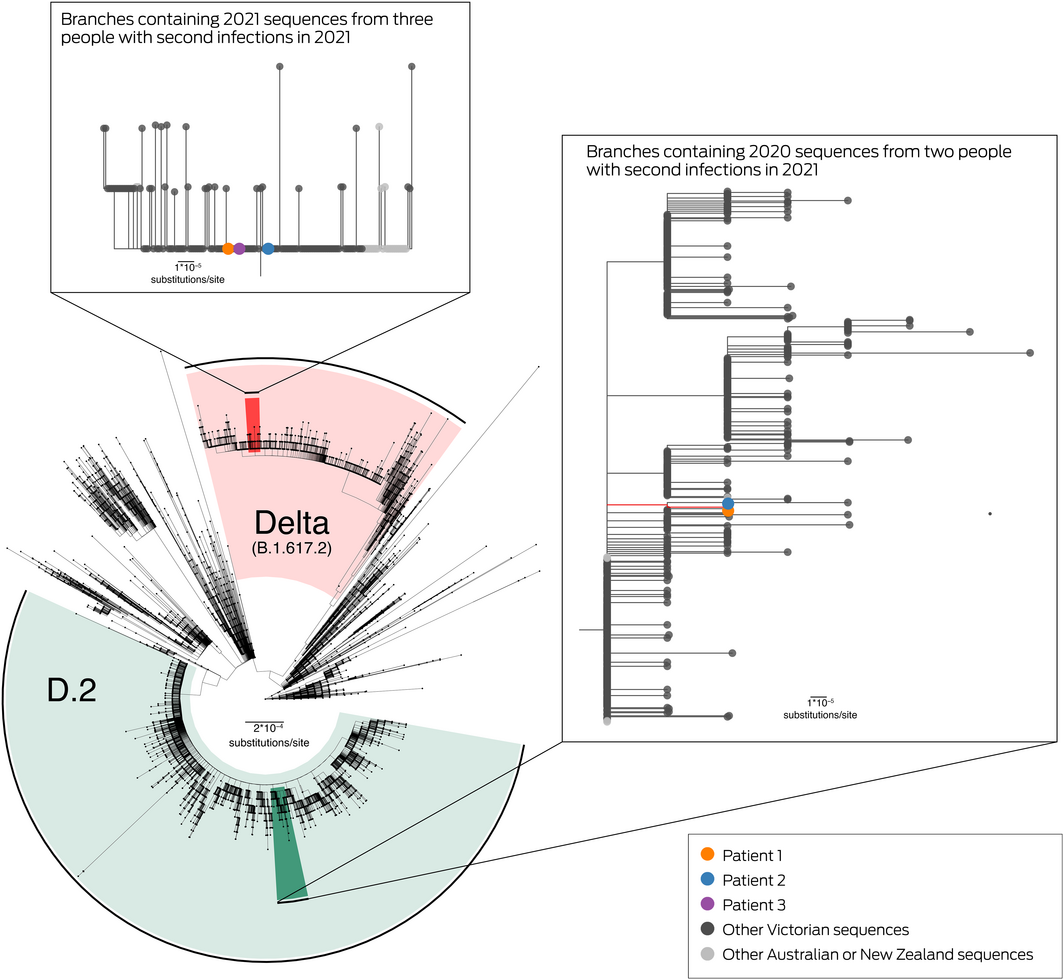
All Victorian SARS‐CoV‐2‐positive samples are required to be referred to the Microbiological Diagnostic Unit Public Health Laboratory in Melbourne for whole genome sequencing and phylogenetic analysis.3 Sequences were available for the three 2021 infections, and for the 2020 infections of patients 1 and 2 (Supporting Information, table 1). The 2021 sequences were genetically distinct from the 2020 sequences, and were closely related to sequences associated with the workplace of patients 1 and 2 and other recent sequences (since June 2021) from Victoria and New South Wales in the Communicable Diseases Genomics Network AusTrakka database (https://www.cdgn.org.au/austrakka) (Box 2; Supporting Information, table 2).
The 2021 sequences were identified as belonging to the SARS‐CoV‐2 variant of concern Delta (pangolin lineage B.1.617.2), first detected overseas in July 2020 and in Australia in February 2021.4 The 2020 sequences were from the pangolin lineage D.2 (not a variant of concern); they were closely related to other 2020 Victorian sequences in the genomic clade dominant during the Victorian “second wave” of June–October 2020, introduced into Victoria following a hotel quarantine breach in May 2020.3
The phylogenetic data, together with the link with a known COVID‐19 outbreak, indicate that the 2021 COVID‐19 diagnoses reflected new infections rather than prolonged viral shedding. Serological data were available only for patients 1 and 3 in 2021; both were seropositive for SARS‐CoV‐2 (Supporting Information, table 2). The three patients had no known immunocompromising conditions and were not eligible for COVID‐19 vaccination at the time of either of their infections. Illness severity was similar during both infections; neither respiratory support nor hospitalisation were required.
People who have recovered from COVID‐19 may be less likely to seek vaccination.5 The occurrence of second infections in Australia, where the incidence of COVID‐19 has been relatively low, indicates that doctors should encourage recovered patients to be vaccinated, and that public awareness of the possibility of reinfection should be promoted to encourage vaccination, testing, and protective behaviours.
Ethics approval
The reported data were collected in accordance with the Victorian Public Health and Wellbeing Act 2008, and the University of Melbourne Human Research Ethics Committee approved the study (study number 1954615.4). The three patients provided written consent for the use of their data in this publication.
Data sharing
All SARS‐CoV‐2 sequence data have been uploaded to the Global Initiative on Sharing All Influenza Data (GISAID; https://www.gisaid.org); the sequence numbers are included in the Supporting Information, table 1.
The metadata for all sequences included in our analysis are provided in the Supporting Information Supporting Information, file 2.
Box 1 – Demographic, sample, and clinical details for three people with second SARS‐CoV‐2 infections, Victoria, 2021
|
Patient |
2020 infections |
2021 infections |
|||||||||||||
|
Number |
Sex |
Age group (years)* |
Symptom onset |
Sample collection date (CT) |
Reported symptoms |
Pangolin lineage |
Symptom onset |
Sample collection date (CT) |
Reported symptoms |
Pangolin lineage |
|||||
|
|
|||||||||||||||
|
1 |
F |
20‒29 |
13 July |
21 July |
Mild† |
D.2 |
16 July |
19 July |
Mild† |
B.1.617.2 |
|||||
|
2 |
M |
20‒29 |
— |
21 July |
None |
D.2 |
20 July |
22 July |
Mild† |
B.1.617.2 |
|||||
|
3 |
M |
20‒29 |
18 July |
22 July |
Mild† |
NA |
16 July |
20 July |
Mild† |
B.1.617.2 |
|||||
|
|
|||||||||||||||
|
CT = polymerase chain reaction cycle threshold (E‐gene target, in‐house assay); NA = not available; SARS‐CoV‐2 = severe acute respiratory syndrome coronavirus 2. * Age group at initial infection. † Cough, runny nose, sore throat, fatigue, headache. |
|||||||||||||||
Box 2 – Maximum likelihood phylogenetic tree of SARS‐CoV‐2 sequences from Australia and New Zealand and for three people with second SARS‐CoV‐2 infections, Victoria, 2021*
SARS‐CoV‐2 = severe acute respiratory syndrome coronavirus 2.
*The phylogenetic tree, prepared using the ggtree package (version 2.4.1) in R 4.0.4, Includes all available sequences from Victoria and all publicly available sequences from other Australian states and New Zealand, to 6 August 2021. Sequences with less than 95% genome coverage are excluded. Highlighted areas indicate sequences identified as variant of concern Delta (red) or D.2 (green) pangolin lineages. Branches containing sequences from patients 1, 2 or 3 are expanded in the insets, and the tree tips are coloured by sequence source. Median pairwise genetic distances are provided in the Supporting Information, table 3.
This figure is available in high‐resolution at https://github.com/MDU‐PHL/COVID19‐paper/blob/master/reinfection/Box2_HighResolution.pdf
Received 13 August 2021, accepted 4 November 2021
- 1. Hansen CH, Michlmayr D, Gubbels SM, et al. Assessment of protection against reinfection with SARS‐CoV‐2 among 4 million PCR‐tested individuals in Denmark in 2020: a population‐level observational study. Lancet 2021; 397: 1204–1212.
- 2. Tillett RL, Sevinsky JR, Hartley PD, et al. Genomic evidence for reinfection with SARS‐CoV‐2: a case study. Lancet Infect Dis 2021; 21: 52–58.
- 3. Lane CR, Sherry NL, Porter AF, et al. Genomics‐informed responses in the elimination of COVID‐19 in Victoria, Australia: an observational, genomic epidemiological study. Lancet Public Health 2021; 6: e547–e556.
- 4. Andersson P, Sherry NL, Howden BP. Surveillance for SARS‐CoV‐2 variants of concern in the Australian context. Medical Journal of Australia 2021; 214: 500–502.e1. https://www.mja.com.au/journal/2021/214/11/surveillance‐sars‐cov‐2‐variants‐concern‐australian‐context
- 5. Hall VJ, Foulkes S, Saei A, et al; SIREN Study Group. COVID‐19 vaccine coverage in health‐care workers in England and effectiveness of BNT162b2 mRNA vaccine against infection (SIREN): a prospective, multicentre, cohort study. Lancet 2021; 397: 1725‐1735.






This investigation was funded by the Victorian Government, and by the National Health and Medical Research Council through the Medical Research Future Fund (MRF9200006). We acknowledge and thank Australian SARS‐CoV‐2 diagnostic and sequencing laboratories for their contributions to this research.
No relevant disclosures.