The known: Real‐time polymerase chain reaction testing of oropharyngeal and bilateral deep nasal swabs for SARS‐CoV‐2 is standard of care in Australia.
The new: Testing saliva as well as standard oropharyngeal‐nasal swabs increased case detection by 59%. Positive test concordance for the two specimen types was 35% (19 of 54 SARS‐CoV‐2‐positive people), but only 11% for children under 10 (two of 19). Providing saliva was preferred to an oropharyngeal‐nasal swab by 335 of 444 participants (75%).
The implications: Adding saliva testing to oropharyngeal‐nasal swab testing increased case detection. Saliva may be suitable as a stand‐alone test specimen for people aged 10 years or more.
Australian guidelines recommend collecting a oropharyngeal and bilateral deep nasal swab for real‐time polymerase chain reaction (PCR) detection of severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2).1 Collecting saliva is less invasive and can be easily undertaken by the tested person themselves, reducing viral exposure for health care workers.2 Four meta‐analyses have found that the pooled sensitivity of saliva real‐time PCR testing for SARS‐CoV‐2 is 84–86% of that with upper respiratory swabs.3,4,5,6 In an analysis of data from 12 groups of tested persons, the overall test concordance between saliva and upper respiratory swabs was 92.1% (κ = 0.84; 95% confidence interval, 0.80–0.87).7
Saliva and upper respiratory swab collection and processing methods can affect test reliability.2 Most studies have assessed self‐collected saliva dribble or posterior oropharyngeal saliva; the comparator upper respiratory swab specimen was usually a nasopharyngeal swab. Further factors include differences in transport medium, sample volumes, timing and severity of illness, assay, and comparator swab collection technique.
Upper respiratory swab collection causes discomfort and may discourage presentation for testing.8 Small studies of children with coronavirus disease 2019 (COVID‐19) found that peak saliva test sensitivity (compared with nasopharyngeal swabs) was 53%9 or 80%10 during the week after symptom onset. An American study including 43 participants aged 4–18 years with COVID‐19 found that positive percent agreement was 79.1% for saliva and 88.4% for nasopharyngeal swabs;11 in a Swiss study including 170 children, positive percent agreement was 93.3% for saliva and 84.4% for nasopharyngeal swabs.12 A further three studies have reported similar SARS‐CoV‐2 detection measures in saliva and nasopharyngeal swabs from children.13,14,15 The diagnostic value of saliva testing is promising, but further validation, including by age group, is needed.
In Australia, respiratory clinics have been established in general practices and hospitals to provide free SARS‐CoV‐2 testing to people meeting testing criteria.16 We compared the concordance and acceptability of saliva testing and standard‐of‐care oropharyngeal‐nasal swab testing in general practices and at a children’s hospital.
Methods
We conducted a multicentre diagnostic validation study including participants recruited from three respiratory clinics in Melbourne paediatric and general practices during the second wave of SARS‐CoV‐2 infections in Victoria.17 We adhere to the Standards for Reporting of Diagnostic Accuracy Studies (STARD) 2015 guidelines.18
Patients of any age who met SARS‐CoV‐2 testing criteria were eligible for inclusion after providing informed consent. Each participant provided a saliva specimen and a standard‐of‐care diagnostic oropharyngeal‐nasal swab. We prioritised recruiting people with confirmed SARS‐CoV‐2 infections and close contacts of people with confirmed SARS‐CoV‐2 infections.
The Royal Children’s Hospital, Melbourne
Research nurses recruited participants (21 July – 18 September 2020) and collected the specimens. Saliva was collected from children under five years of age in a SalivaBio swab and storage tube (Stratech Scientific); older participants were asked to dribble at least 2 mL saliva into a collection pot without transport medium. All participants underwent standard‐of‐care oropharyngeal‐nasal swabbing (dry FLOQSwabs, Copan). Specimens were tested in the Royal Children’s Hospital diagnostic molecular microbiology laboratory. Swabs were eluted into 500 µL phosphate‐buffered saline (PBS); saliva specimens (except those collected with the SalivaBio system) were diluted 1:1 in PBS. For each specimen type, nucleic acids in 200 µL preparations were extracted with the MagNA Pure 96 extraction system (Roche). Extracts were tested with the LightMix modular SARS and Wuhan CoV E‐gene kit (TIB Molbiol) in the LightCycler 480 II real‐time PCR system (Roche). The status of E‐gene‐positive specimen extracts was confirmed with the Respiratory Pathogens 16‐well assay (targeting the open reading frames ORF‐1 and ORF‐8) on the High‐Plex 24 unit (AusDiagnostics).
General practices
Participants were recruited at cohealth West Melbourne, a fixed site respiratory clinic, during 27 July – 18 September 2020 and at Cirqit Health Altona North, a general practice respiratory clinic with drive‐through facility, during 18 September – 2 October 2020. Clinic staff recruited participants, undertook standard‐of‐care oropharyngeal‐nasal swab collection, and asked participants to dribble at least 2 mL saliva into collection pots without transport medium. Saliva and oropharyngeal‐nasal swabs were tested for SARS‐CoV‐2 at the Microbiological Diagnostic Unit Public Health Laboratory within 48 hours of collection. Saliva was diluted 1:4 in saline; dry swabs were resuspended in 3 mL saline for testing with the Aptima SARS‐CoV‐2 or Panther Fusion SARS‐CoV‐2 assays (both Hologic).
Study surveys
Participants or their guardians were invited to complete a survey about their symptoms and their specimen collection preferences. At the Royal Children’s Hospital, surveys were administered by research nurses, who also collected exposure risk factor data. For general practice participants, an online REDCap survey was accessed via a QR code (cohealth, West Melbourne) or texted to the participants’ mobile phones (Cirqit Health, Altona North); exposure risk factor data were not collected.
Study definitions
A specimen was deemed positive if at least two SARS‐CoV‐2 gene targets were detected, consistent with national guidelines.1 All positive tests were considered true positives; that is, a participant was deemed to be positive for SARS‐CoV‐2 if test results for either specimen type were positive. Specimens with indeterminate, invalid, or missing results (including saliva specimens with insufficient volume for testing) were deemed non‐assessable and excluded from analysis.
Statistical analysis
Our target sample size was 38 participants with confirmed SARS‐CoV‐2 infections (required precision, 9%; assumed κ = 0.84).19 The positive percent agreement (PPA) was reported as the proportion of all SARS‐CoV‐2‐positive people who had SARS‐CoV‐2 detected in each specimen type (saliva and oropharyngeal‐nasal swab). Concordance between paired specimens for SARS‐CoV‐2 detection is reported as the κ statistic and as overall percentage agreement. Proportions of positive specimens (with exact binomial 95% confidence intervals [CIs]) were calculated for each specimen type and compared in McNemar tests, both overall and by subgroup. Ratios of SARS‐CoV‐2 detections in paired specimens (oropharyngeal‐nasal swab and saliva) to detections in single specimen types (oropharyngeal‐nasal swab or saliva) are also reported (with 95% CIs).
Analyses were undertaken in Stata 16; P < 0.05 was deemed statistically significant.
Ethics approval
The study was approved by the Royal Children’s Hospital Human Research Ethics Committee (HREC/65175/RCHM‐2020).
Results
A total of 1165 participants were recruited, of whom 115 (10%) were excluded because of non‐assessable results; 110 saliva specimens were non‐assessable (9%), including 86 with insufficient volume for testing and four that leaked (Supporting Information, figure).
Participant baseline data
Of the 1050 included participants, 176 were children under 10 years of age (17%) and 749 were recruited from general practices (71%). In 54 cases (5%), SARS‐CoV‐2 was detected in saliva or oropharyngeal‐nasal swabs; their median age was 27.6 years (range, 6 months to 65 years), and 30 were men. Twenty‐two of the 38 SARS‐CoV‐2‐positive people for whom risk factor data were available had been close contacts of people with confirmed SARS‐CoV‐2 infections (58%), and 19 of the 38 for whom symptom information was available reported symptoms at the time of specimen collection, most frequently cough (nine people, 24%) (Box 1). For ten of the 11 SARS‐CoV‐2‐positive people who reported symptoms and for whom the symptom onset date was known, symptom duration was less than four days at the time of specimen collection (median, 2 days; range, 0–12 days).
Positive percent agreement
The overall PPA was 72% (95% CI, 58–84%) for saliva and 63% (95% CI, 49–76%) for oropharyngeal‐nasal swabs. For the 35 positive specimens from people aged 10 years or more, PPA was 86% (95% CI, 70–95%) for saliva and 63% (95% CI, 45–79%) for oropharyngeal‐nasal swabs; for the 19 positive specimens from people under 10 years of age, PPA was 47% (95% CI, 24–71%) for saliva and 63% (95% CI, 38–84%) for oropharyngeal‐nasal swabs (Box 2; Supporting Information, table 1).
Paired and single specimen testing for SARS‐CoV‐2
For 19 of 54 participants with positive SARS‐CoV‐2 test results, both specimens were positive (35%), in 20 cases only the saliva test result was positive (37%), and in 15 cases only the oropharyngeal‐nasal swab test result was positive (28%) (Box 1). Adding a paired saliva specimen to standard‐of‐care oropharyngeal‐nasal swab testing increased total case detection by 59% (95% CI, 29–95%); it increased case detection by 90% (95% CI, 24–191%) in asymptomatic people and by 100% (95% CI, 29–210%) in females. Adding a paired oropharyngeal‐nasal swab to a single saliva specimen increased overall total case detection by 38% (95% CI, 17–63%), but only by 17% (95% CI, 2–34%) in people aged 10 years or more (Box 3).
Children positive for SARS‐CoV‐2
Seven of the 19 children under 10 years of age positive for SARS‐CoV‐2 were symptomatic at the time of specimen collection, including four of nine with positive saliva specimen test results and four of 12 with positive oropharyngeal‐nasal swab test results. Test results were positive for both specimen types for two children (Box 1). Total case detection in children under 10 years of age was increased by 58% (95% CI, 12–123%) by adding a paired saliva specimen to the standard‐of‐care oropharyngeal‐nasal swab, and by 111% (95% CI, 31–239%) by adding a paired oropharyngeal‐nasal swab to the saliva specimen for testing (Box 3). Of the 21 SARS‐CoV‐2‐positive people under 18 years of age, both specimens were positive in two cases, only the saliva specimen in eight, and only the oropharyngeal‐nasal swab in 11 (Box 1). SARS‐CoV‐2 was detected in oropharyngeal‐nasal swabs from all four SARS‐CoV‐2‐positive infants (under 12 months of age), but in only one saliva specimen.
Test concordance
Overall, 1015 of 1050 paired test results were concordant (97%; κ = 0.50; 95% CI, 0.36–0.65). Specifically, 159 of 176 test results for children under 10 years of age were concordant (90%; κ = 0.14; 95% CI, ‒0.09 to 0.37), and 856 of 874 test results for people aged 10 years or more (98%; κ = 0.64; 95% CI, 0.49–0.80).
Specimen preference
Saliva collection was preferred to an oropharyngeal‐nasal swab by 335 of 444 participants who provided a response to the survey question (75%), including 141 of 153 children under 10 years of age (92%) (Box 4).
Discussion
Accurately identifying people with SARS‐CoV‐2 infections is essential for case management and preventing transmission. We found that testing saliva as well as the standard‐of‐care oropharyngeal‐nasal swab increased case detection by 59%. The overall PPA for saliva testing was similar to that for oropharyngeal‐nasal swabs, but saliva testing alone would have missed ten of 19 infections (53%) in children under 10 years of age.
Three in four people preferred providing saliva to undergoing oropharyngeal‐nasal swabbing, including more than 90% of children under 10 years of age. Saliva collection was considered more comfortable, more convenient, and preferable to self‐collected nasopharyngeal swabs in a study in Singapore.20 An online discrete choice experiment survey of 4793 adults found that nasopharyngeal swabbing may deter people from being tested, a problem that could be mitigated by using saliva specimens.21 Saliva testing may be particularly attractive for people unwilling or unable to undergo oropharyngeal‐nasal swabbing and in settings where repeat testing is desired, such as higher risk workplaces.
We estimated the PPAs for oropharyngeal‐nasal swabs and saliva as proxy measures of test sensitivity. Positive test result concordance in our study was low (35%), and the PPAs for saliva and upper respiratory swabs were lower than in many other studies. Our comparator specimen was the oropharyngeal‐nasal swab, consistent with national guidelines,22 rather than the nasopharyngeal swab used in many studies. Our lower PPA for oropharyngeal‐nasal swabs might reflect lower test sensitivity than with nasopharyngeal swabs; however, one meta‐analysis found that test sensitivity with pooled throat and nasal swabs was 97% (95% CI, 93–100%) of that with nasopharyngeal swabs.6 Positive test result concordance was very low for people under 18 years of age (two of 21, 10%), but the value for adults (17 of 33, 52%) was similar to that in a Canadian study (70 adults) in which saliva and oropharyngeal‐nasal swabs or nasopharyngeal swabs were tested (49%),23 but lower than a British study in which oropharyngeal‐nasal swabs were the comparator (74%).24
In addition to the comparator specimen used, our study differed from similar studies with respect to testing children. In a Singapore study of 18 children diagnosed with COVID‐19 (using nasopharyngeal swabs), peak sensitivity of saliva testing (52.9%) was on days 4–7;9 in a Korean study of 11 children diagnosed with COVID‐19 (using nasopharyngeal swabs), overall sensitivity of saliva testing was 73%, but fell from 80% (week 1) to 11% (week 3).10 The denominators for the sensitivity calculations in both these studies comprised children with SARS‐CoV‐2‐positive nasopharyngeal swabs, assumed to be the gold standard. As our study included 176 children under 10 years of age (and 212 people under 18 years) with suspected COVID‐19, we could more fully explore the role of saliva testing for detecting SARS‐CoV‐2. We included cases in our PPA denominator for which positive test results were obtained with either specimen, recognising that there is no gold standard test specimen. We would have missed 37% of SARS‐CoV‐2‐positive children under 10 had paired saliva specimens not been added to the oropharyngeal‐nasal swab for testing, consistent with other recent studies in children.13,14
Several studies with nasopharyngeal swabs as the comparator have reported higher PPAs for children than ours for saliva (80.0–93.9%;11,12,15 our study, 47%) and upper respiratory swab testing (86.7%;11 our study, 63%). Our study included 14 children under four years of age, while nearly all participants in other investigations were over four years old. We found that testing oropharyngeal‐nasal swabs as well as saliva more than doubled case detection in children under 10 years of age. Our findings suggest that saliva testing of children with suspected infections is most appropriate as an adjunct to rather than as a replacement for standard‐of‐care oropharyngeal‐nasal swabs. Adding an oropharyngeal‐nasal swab to saliva testing increased case detection in people over 10 years of age by only 17%, suggesting that saliva testing could be substituted for oropharyngeal‐nasal swab testing in older children and adults.
Limitations
An important limitation was that 9% of saliva specimens were non‐assessable, most frequently because the volume was inadequate. Another study found that at least one‐third of pure saliva specimens were difficult to pipette.25 A much lower invalid specimen rate (0.03%) pertained when saliva was collected with a straw‐like device, followed by centrifugation and the addition of proteinase K.26 Other options to reduce the invalid specimen rate could include participant education, marking the required volume on the collection pot, and using alternative collection devices.27 When saliva volume was adequate for testing, the higher SARS‐CoV‐2 detection rate than with oropharyngeal‐nasal swabs may have been related to the larger specimen volume.
We undertook a rapid real life study, assessing saliva collection and testing in symptomatic and asymptomatic people in several settings, including a drive‐through testing centre. In settings of low incidence, targeted sampling may be required to increase the number of SARS‐CoV‐2‐positive cases and to increase study power. As the local incidence of COVID‐19 was declining rapidly during the recruitment period, we focused on people known to be SARS‐CoV‐2‐positive and their close contacts to achieve our target sample size. We could not synchronise procedures and recruitment across sites because of staff workloads and the embedded nature of specimen processing. Real‐time PCR analysis was undertaken in two laboratories using different assay platforms. Results from assays using differing detection parameters were pooled. We assumed that all positive results were true positives; that is, we did not allow for potentially false positive results. At the Royal Children’s Hospital, we used a different saliva collection method for children under 5 years of age, limiting the comparability between study sites of findings for this age group.
Conclusion
Testing paired saliva and oropharyngeal‐nasal swab specimens for SARS‐CoV‐2 increases the detection of infections, particularly in children under 10 years of age. In older children and adults, testing saliva alone may be appropriate given the general preference of people for saliva collection to oropharyngeal‐nasal swabbing and the adequacy of SARS‐CoV‐2 detection in saliva.
Box 1 – SARS‐CoV‐2 testing results for 1050 participants, by characteristic and specimen type (saliva or oropharyngeal and bilateral deep nasal swab)
|
Characteristic |
|
SARS‐CoV‐2 test result |
Positive results, by specimen type |
||||||||||||
|
Participants |
Not detected |
Detected |
Saliva |
Saliva only |
Swab only |
||||||||||
|
|
|||||||||||||||
|
Number of participants |
1050 |
996 (95%) |
54 (5%) |
19 (35%) |
20 (37%) |
15 (28%) |
|||||||||
|
Laboratory site |
|
|
|
|
|
|
|||||||||
|
Royal Children’s Hospital |
301 (29%) |
261 (87%) |
40 (13%) |
11 (28%) |
17 (43%) |
12 (30%) |
|||||||||
|
Primary care sites |
749 (71%) |
735 (98%) |
14 (2%) |
8 (57%) |
3 (21%) |
3 (21%) |
|||||||||
|
Fixed site |
575 (77%) |
563 (98%) |
12 (2%) |
7 (58%) |
2 (17%) |
3 (25%) |
|||||||||
|
Drive‐through |
174 (23%) |
172 (99%) |
2 (1%) |
1 (50%) |
1 (50%) |
0 |
|||||||||
|
Age group (years) |
|
|
|
|
|
|
|||||||||
|
< 5 |
129 (12%) |
114 (88%) |
15 (12%) |
2 (13%) |
5 (33%) |
8 (53%) |
|||||||||
|
5–9 |
47 (5%) |
43 (92%) |
4 (9%) |
0 |
2 (50%) |
2 (50%) |
|||||||||
|
10–17 |
36 (3%) |
34 (94%) |
2 (6%) |
0 |
1 (50%) |
1 (50%) |
|||||||||
|
≥ 18 |
838 (80%) |
805 (96%) |
33 (4%) |
17 (52%) |
12 (36%) |
4 (12%) |
|||||||||
|
Sex |
|
|
|
|
|
|
|||||||||
|
Male |
375 (36%) |
345 (92%) |
30 (8%) |
12 (40%) |
9 (30%) |
9 (30%) |
|||||||||
|
Female |
432 (41%) |
412 (95%) |
20 (5%) |
4 (20%) |
10 (50%) |
6 (30%) |
|||||||||
|
Unknown |
243 (23%) |
239 (98%) |
4 (2%) |
3 (75%) |
1 (25%) |
0 |
|||||||||
|
Risk factors (N = 298*) |
|
|
|
|
|
|
|||||||||
|
Close contact |
168 (56%) |
146 (87%) |
22 (13%) |
3 (14%) |
10 (46%) |
9 (41%) |
|||||||||
|
Health care worker in household |
12 (4%) |
9 (75%) |
3 (25%) |
0 |
2 (67%) |
1 (33%) |
|||||||||
|
Recent positive test |
22 (7%) |
12 (55%) |
10 (46%) |
6 (60%) |
1 (10%) |
3 (30%) |
|||||||||
|
No identified risk factor |
107 (36%) |
101 (94%) |
6 (6%) |
0 |
6 (100%) |
0 |
|||||||||
|
Symptoms† (N = 465) |
|
|
|
|
|
|
|||||||||
|
No |
199 (43%) |
180 (91%) |
19 (10%) |
1 (5%) |
9 (47%) |
9 (47%) |
|||||||||
|
Yes (any) |
266 (57%) |
247 (93%) |
19 (7%) |
8 (42%) |
8 (42%) |
3 (16%) |
|||||||||
|
Sore throat |
107 (23%) |
101 (94%) |
6 (6%) |
3 (50%) |
2 (33%) |
1 (17%) |
|||||||||
|
Cough |
103 (22%) |
94 (91%) |
9 (9%) |
6 (67%) |
2 (22%) |
1 (11%) |
|||||||||
|
Fatigue |
55 (12%) |
51 (93%) |
4 (7%) |
1 (25%) |
2 (50%) |
1 (25%) |
|||||||||
|
Fever |
53 (11%) |
47 (89%) |
6 (11%) |
3 (50%) |
2 (33%) |
1 (17%) |
|||||||||
|
Runny nose (N = 298‡) |
93 (31%) |
88 (95%) |
5 (5%) |
2 (40%) |
3 (60%) |
0 |
|||||||||
|
Headache (N = 298‡) |
30 (10%) |
26 (87%) |
4 (13%) |
3 (75%) |
1 (25%) |
0 |
|||||||||
|
|
|||||||||||||||
|
* Royal Children’s Hospital participants assessed only, none of whom reported recent international travel. † Less frequent symptoms: myalgia, 31 (6%); diarrhoea, 15 (3%); dyspnoea, 12 (2%); anosmia, four (1%); confusion, three (1%); dysgeusia, two (< 0.1%). ‡ Royal Children’s Hospital participants assessed only. |
|||||||||||||||
Box 2 – Positive percent agreement (PPA) for saliva and oropharyngeal and bilateral deep nasal swab testing for SARS‐CoV‐2, for 54 SARS‐CoV‐2‐positive participants
|
Characteristic |
Positive specimens |
Saliva specimens |
Swab specimens |
P* |
|||||||||||
|
Number |
PPA (95% CI) |
Number |
PPA (95% CI) |
||||||||||||
|
|
|||||||||||||||
|
All positive specimens |
54 |
39 |
72% (58–84%) |
34 |
63% (49–76%) |
0.40 |
|||||||||
|
Age group (years) |
|
|
|
|
|
|
|||||||||
|
Under 10 |
19 |
9 |
47% (24–71%) |
12 |
63% (38–84%) |
0.47 |
|||||||||
|
10 or more |
35 |
30 |
86% (70–95%) |
22 |
63% (45–79%) |
0.06 |
|||||||||
|
Sex |
|
|
|
|
|
|
|||||||||
|
Male |
30 |
21 |
70% (51–85%) |
21 |
70% (51–85%) |
1.0 |
|||||||||
|
Female |
20 |
14 |
70% (46–88%) |
10 |
50% (27–73%) |
0.32 |
|||||||||
|
Unknown |
4 |
4 |
100% (40–100%)† |
3 |
75% (19–99%) |
0.32 |
|||||||||
|
Laboratory |
|
|
|
|
|
|
|||||||||
|
Royal Children’s Hospital |
40 |
28 |
70% (53–83%) |
23 |
58% (41–73%) |
0.35 |
|||||||||
|
Microbiological Diagnostic Unit |
14 |
11 |
79% (49–95%) |
11 |
79% (49–95%) |
1.0 |
|||||||||
|
Symptoms at time of test |
|
|
|
|
|
|
|||||||||
|
Symptomatic |
19 |
16 |
84% (60–97%) |
11 |
58% (33–80%) |
0.13 |
|||||||||
|
Asymptomatic |
19 |
10 |
53% (29–76%) |
10 |
53% (29–76%) |
1.0 |
|||||||||
|
Unknown |
16 |
13 |
81% (54–96%) |
13 |
81% (54–96%) |
1.0 |
|||||||||
|
|
|||||||||||||||
|
* For difference in positive proportions between saliva and swab test results (McNemar test). † One‐sided 95% confidence interval. |
|||||||||||||||
Box 3 – Ratio of SARS‐CoV‐2 detections in paired saliva and oropharyngeal and bilateral deep nasal swab specimens to detections in single specimen types, with 95% confidence intervals
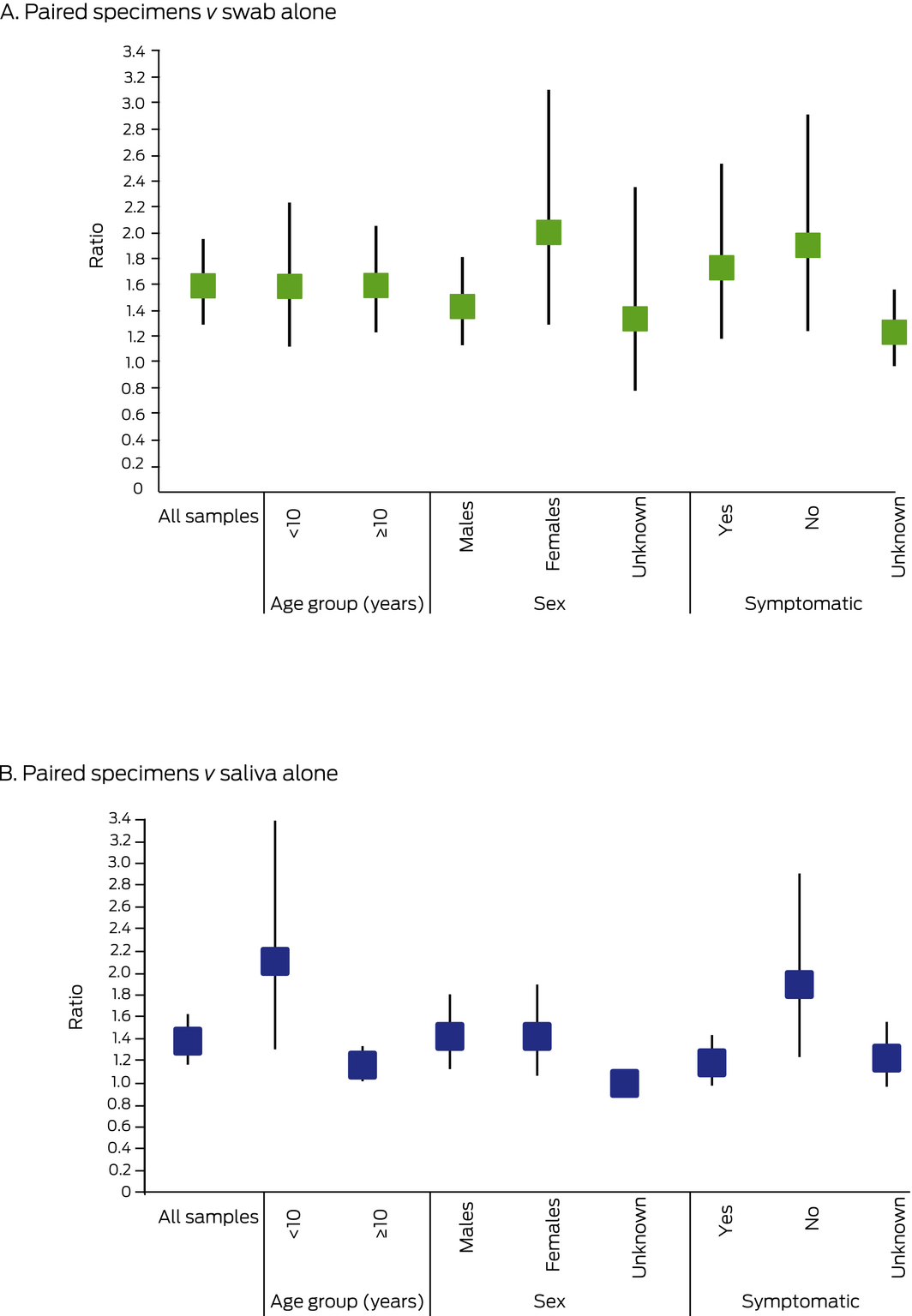
The raw data for this graph is included in the Supporting Information, table 3.
Box 4 – Participant preference regarding specimen collection type (saliva or oropharyngeal and bilateral deep nasal swab)
|
Characteristic |
Responses |
No preference |
Swab preferred |
Saliva preferred |
|||||||||||
|
|
|||||||||||||||
|
All participants |
444 |
69 (16%) |
40 (9%) |
335 (75%) |
|||||||||||
|
Age group (years) |
|
|
|
|
|||||||||||
|
< 5 |
110 |
6 (6%) |
4 (4%) |
100 (91%) |
|||||||||||
|
5–9 |
43 |
0 |
2 (5%) |
41 (95%) |
|||||||||||
|
10–17 |
25 |
4 (16%) |
3 (12%) |
18 (72%) |
|||||||||||
|
≥ 18 |
266 |
59 (22%) |
31 (12%) |
176 (66%) |
|||||||||||
|
Sex |
|
|
|
|
|||||||||||
|
Male |
181 |
21 (12%) |
12 (7%) |
148 (82%) |
|||||||||||
|
Female |
223 |
32 (14%) |
22 (10%) |
169 (76%) |
|||||||||||
|
Unknown |
40 |
16 (40%) |
6 (15%) |
18 (45%) |
|||||||||||
|
|
|||||||||||||||
|
|
|||||||||||||||
Received 28 February 2026, accepted 28 February 2026
- Jane Oliver1
- Shidan Tosif2
- Lai‐yang Lee2
- Anna‐Maria Costa2
- Chelsea Bartel2
- Katherine Last2
- Vanessa Clifford2,3
- Andrew Daley2,4
- Nicole Allard1,5
- Catherine Orr5
- Ashley Nind5
- Karyn Alexander3,6
- Niamh Meagher7
- Michelle Sait8
- Susan A Ballard8
- Eloise Williams9
- Katherine Bond10
- Deborah A Williamson8,11
- Nigel W Crawford12
- Katherine B Gibney1,9
- 1 The Peter Doherty Institute for Infection and Immunity, University of Melbourne, Melbourne, VIC
- 2 The Royal Children’s Hospital, Melbourne, VIC
- 3 Melbourne Medical School, University of Melbourne, Melbourne, VIC
- 4 The Royal Women’s Hospital, Melbourne, VIC
- 5 cohealth, Melbourne, VIC
- 6 Cirqit Health, Melbourne, VIC
- 7 The University of Melbourne, Melbourne, VIC
- 8 Public Health Laboratory, University of Melbourne, Melbourne, VIC
- 9 Royal Melbourne Hospital, Melbourne, VIC
- 10 Victorian Infectious Diseases Reference Laboratory, Melbourne Health, Melbourne, VIC
- 11 Melbourne Health, Melbourne, VIC
- 12 Surveillance of Adverse Events Following Vaccination in the Community (SAEFVIC), Murdoch Children’s Research Institute, Melbourne, VIC
Our study was supported by a donation from the Isabel and John Gilbertson Charitable Trust. We acknowledge all participants, and the clinical, administrative and laboratory staff who assisted our study at the Royal Children’s Hospital Melbourne, cohealth, Cirqit Health, the Microbiological Diagnostic Unit Public Health Laboratory, Golden Messenger, the Royal Melbourne Hospital, and the University of Melbourne.
No relevant disclosures.
- 1. Public Health Laboratory Network. PHLN guidance on laboratory testing for SARS‐CoV‐2 (the virus that causes COVID‐19), version 1.16. Updated Feb 2021. https://www.health.gov.au/resources/publications/phln-guidance-on-laboratory-testing-for-sars-cov-2-the-virus-that-causes-covid-19 (viewed May 2021).
- 2. Fernandes LL, Pacheco VB, Borges L, et al. Saliva in the diagnosis of COVID‐19: a review and new research directions. J Dent Res 2020; 99: 1435–1443.
- 3. Kivelä JM, Jarva H, Lappalainen M, Kurkela S. Saliva‐based testing for diagnosis of SARS‐CoV‐2 infection: a meta‐analysis. J Med Virol 2021; 93: 1256–1258.
- 4. Khiabani K, Amirzade‐Iranaq MH. Are saliva and deep throat sputum as reliable as common respiratory specimens for SARS‐CoV‐2 detection? A systematic review and meta‐analysis. Am J Infect Control 2021; https://doi.org/10.1016/j.ajic.2021.03.008 [online ahead of print].
- 5. Buban JM, Villanueva PN, Gregorio GEV. Should RT‐PCR of saliva samples be used for diagnosis of COVID‐19? (Philippine COVID‐19 Living Clinical Practice Guidelines). Updated 15 Mar 2021. https://www.psmid.org/wp-content/uploads/2021/05/SALIVA-RT-PCR-CPG-FINAL_031521_MMA.pdf (viewed May 2021).
- 6. Tsang NNY, So HC, Ng KY, et al. Diagnostic performance of different sampling approaches for SARS‐CoV‐2 RT‐PCR testing: a systematic review and meta‐analysis. Lancet Infect Dis 2021; https://doi.org/10.1016/S1473-3099(21)00146-8 [online ahead of print].
- 7. Zhu J, Guo J, Xu Y, Chen X. Viral dynamics of SARS‐CoV‐2 in saliva from infected patients. J Infect 2020; 81: e48–e50.
- 8. Ruggiero A, Sanguinetti M, Gatto A, et al. Diagnosis of COVID‐19 infection in children: less nasopharyngeal swabs, more saliva. Acta Paediatr 2020; 109: 1913–1914.
- 9. Chong CY, Kam KQ, Li J, et al. Saliva is not a useful diagnostic specimen in children with coronavirus disease 2019 [letter]. Clin Infect Dis 2020; https://doi.org/10.1093/cid/ciaa1376 [online ahead of print].
- 10. Han MS, Seong MW, Kim N, et al. Viral RNA load in mildly symptomatic and asymptomatic children with COVID‐19, Seoul, South Korea. Emerg Infect Dis 2020; 26: 2497–2499.
- 11. Yee R, Truong TT, Pannaraj PS, et al. Saliva is a promising alternative specimen for the detection of SARS‐CoV‐2 in children and adults. J Clin Microbiol 2021; 59: e02686–20.
- 12. Huber M, Schreiber PW, Scheier T, et al. High efficacy of saliva in detecting SARS‐CoV‐2 by RT‐PCR in adults and children. Microorganisms 2021; 9: 642.
- 13. Fougère Y, Schwob JM, Miauton A, et al. Performance of RT‐PCR on saliva specimens compared to nasopharyngeal swabs for the detection of SARS‐CoV‐2 in children: a prospective comparative clinical trial [preprint]. medRxiv , 1 Mar 2021. https://doi.org/10.1101/2021.02.27.21252571 (viewed May 2021).
- 14. Felix AC, De Paula AV, Ribeiro AC, et al. Saliva as a reliable sample for COVID‐19 diagnosis in paediatric patients [preprint]. medRxiv , 31 Mar 2021. https://doi.org/10.1101/2021.03.29.21254566 (viewed May 2021).
- 15. Al Suwaidi H, Senok A, Varghese R, et al. Saliva for molecular detection of SARS‐CoV‐2 in school‐age children. Clin Microbiol Infect 2021; https://doi.org/10.1016/j.cmi.2021.02.009 [online ahead of print].
- 16. Department of Health and Human Services (Victoria). Assessment and testing criteria for coronavirus (COVID‐19). Updated 9 Apr 2021. https://www.dhhs.vic.gov.au/assessment-and-testing-criteria-coronavirus-covid-19 (viewed May 2021).
- 17. Department of Health and Human Services (Victoria). Coronavirus (COVID‐19). https://www.dhhs.vic.gov.au/coronavirus (viewed May 2021).
- 18. Cohen JF, Korevaar DA, Altman DG, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 2016; 6: e012799.
- 19. Williams E, Bond K, Zhang B, et al. Saliva as a noninvasive specimen for detection of SARS‐CoV‐2. J Clin Microbiol 2020; 58: e00776–20.
- 20. Ku CW, Shivani D, Kwan JQT, et al. Validation of self‐collected buccal swab and saliva as a diagnostic tool for COVID‐19. Int J Infect Dis 2021; 104: 255–261.
- 21. Zimba R, Kulkarni S, Berry A, et al; the CHASING COVID Cohort Study Team. Testing, testing: what SARS‐CoV‐2 testing services do adults in the United States actually want? [preprint]. medRxiv, 18 Sept 2020. https://doi.org/10.1101/2020.09.15.20195180 (viewed May 2021).
- 22. Communicable Diseases Network Australia. Coronavirus disease 2019 (COVID‐19): CDNA national guidelines for public health units; version 3.1. Updated 4 June 2020. https://www1.health.gov.au/internet/main/publishing.nsf/Content/cdna-song-novel-coronavirus.htm (viewed May 2021).
- 23. Caulley L, Corsten M, Eapen L, et al. Salivary detection of COVID‐19. Ann Intern Med 2021; 174: 131–133.
- 24. Byrne RL, Kay GA, Kontogianni K, et al. Saliva offers a sensitive, specific and non‐invasive alternative to upper respiratory swabs for SARS‐CoV‐2 diagnosis [pre‐print]. medRxiv, 11 July 2020. https://doi.org/10.1101/2020.07.09.20149534 (viewed May 2021).
- 25. Landry ML, Criscuolo J, Peaper DR. Challenges in use of saliva for detection of SARS CoV‐2 RNA in symptomatic outpatients. J Clin Virol 2020; 130: 104567.
- 26. Vogels CBF, Watkins AE, Harden CA, et al. SalivaDirect: a simplified and flexible platform to enhance SARS‐CoV‐2 testing capacity. Med (NY) 2021; 2: 263–280.
- 27. Ceron JJ, Lamy E, Martinez‐Subiela S,et al. Use of saliva for diagnosis and monitoring the SARS‐CoV‐2: a general perspective. J Clin Med 2020; 9: 1491.






Abstract
Objective: To compare the concordance and acceptability of saliva testing with standard‐of‐care oropharyngeal and bilateral deep nasal swab testing for severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) in children and in general practice.
Design: Prospective multicentre diagnostic validation study.
Setting: Royal Children’s Hospital, and two general practices (cohealth, West Melbourne; Cirqit Health, Altona North) in Melbourne, July–October 2020.
Participants: 1050 people who provided paired saliva and oropharyngeal‐nasal swabs for SARS‐CoV‐2 testing.
Main outcome measures: Numbers of cases in which SARS‐CoV‐2 was detected in either specimen type by real‐time polymerase chain reaction; concordance of results for paired specimens; positive percent agreement (PPA) for virus detection, by specimen type.
Results: SARS‐CoV‐2 was detected in 54 of 1050 people with assessable specimens (5%), including 19 cases (35%) in which both specimens were positive. The overall PPA was 72% (95% CI, 58–84%) for saliva and 63% (95% CI, 49–76%) for oropharyngeal‐nasal swabs. For the 35 positive specimens from people aged 10 years or more, PPA was 86% (95% CI, 70–95%) for saliva and 63% (95% CI, 45–79%) for oropharyngeal‐nasal swabs. Adding saliva testing to standard‐of‐care oropharyngeal‐nasal swab testing increased overall case detection by 59% (95% CI, 29–95%). Providing saliva was preferred to an oropharyngeal‐nasal swab by most participants (75%), including 141 of 153 children under 10 years of age (92%).
Conclusion: In children over 10 years of age and adults, saliva testing alone may be suitable for SARS‐CoV‐2 detection, while for children under 10, saliva testing may be suitable as an adjunct to oropharyngeal‐nasal swab testing for increasing case detection.