Benefiting from genomics in health care depends on data sharing and digital health integration
The knowledge gained from patients’ genome sequences will increasingly help inform their diagnoses and clinical management. For the health system, the aims of genomic medicine are faster and more accurate diagnosis, and the correct treatment first time. Research has already demonstrated reduced overall cost when genome sequencing is used for diagnostic purposes.1 For patients, genomic medicine offers personalised care, empowerment, access to information and control of the diagnostic odyssey. In some clinical areas, genomics has already been shown to provide faster and more accurate disease diagnosis; identify the most effective treatment for cancer; and guide pharmaceutical use with information such as risk of adverse events. As we learn more, considerable value will also come from using genomics to understand predisposition to disease, enable early detection of disease, and optimise disease management and prevention strategies.
The sequencing of the human genome in 2000 was expected to catalyse a revolution in the treatment of disease. However, the use of genomics in a systematic way is only now dawning in the Australian health care system. In the next decade, knowledge about how variation in our genomes affects our health is expected to increase dramatically. So we now face the technical, ethical and social challenges of how health services should use genomic information to improve health care for all Australians. The Australian Genomics Health Alliance (hereafter referred to as Australian Genomics) is a National Health and Medical Research Council‐funded initiative that includes over 400 investigators and collaborators, across 100 sites nationally, and undertakes research into the integration of genomics into clinical care. Australian Genomics collaborates with other Australian genomic initiatives, including the Melbourne Genomics Health Alliance2, Sydney Genomics Collaborative and Queensland Genomics Health Alliance, and internationally through the Global Alliance for Genomics and Health.3
Maximising the benefits of genomic information requires comprehensive, targeted clinical data — to inform the use of genomic sequences and to optimise interrogation of the relationship between a genotype and a clinical phenotype. This is where high quality data captured in electronic medical records, and the national My Health Record system, will become increasingly important. These systems provide opportunities for the health care system and patients to use genomic information to inform health care decisions throughout life.
Australian Genomics: towards national data registries and standards for genomic medicine (Program 2)
Australian Genomics is undertaking a series of clinical flagship projects across cancer and rare diseases to identify the opportunities, challenges and bottlenecks in integrating genomics into clinical care. The program is underpinned by four research programs: diagnostic approaches; national data infrastructure and services; implementation, economics, policy and ethics; and workforce and education. One of these projects — A National Approach to Data Federation and Analysis (Program 2) — is shaping a coordinated, national approach to generating, processing, curating, storing and sharing genomic and related clinical data. Systems and policies that align with international best practice are being piloted.2
This program has identified several key challenges in standardising and sharing genomic data for clinical use:
-
establishing database systems that link genomic data with phenotype and clinical characteristics — commonly known as Gen‐Phen databases
-
accessing the required computing power and large scale data storage for federated data analysis and dissemination
-
manually curating sequence data once it is processed by bioinformatics pipelines
-
linking genomic information to health records; and
-
managing large collections of sequence data over time.
Linking genotype and phenotype
A key challenge for the use of genomics in clinical care is the delivery of high quality information about the patient's genome that may inform the clinician about the diagnosis, prognosis or treatment decision in the context of the patient's clinical presentation. Gen‐Phen databases need to use standardised machine‐readable terminology for clinical phenotype to support digital integration into the health data ecosystem. Standardised clinical terminology, such as SNOMED CT (Systematized Nomenclature of Medicine – Clinical Terms)4 and Human Phenotype Ontology (HPO),5 are increasingly being used to describe diagnoses and symptoms in electronic medical records, and can be used computationally to describe an individual's clinical phenotype.
Storing and sharing genomic data
The genotype is represented by variations in a patients’ sequence when compared with a reference human genome.6 Bioinformatics pipelines use considerable computing power to process and analyse sequence data from a sequencing machine. In doing so, an individual's sequence data are aligned against the reference genome, so that differences (known as variants) can be identified.
A major challenge for the Australian health care system will be establishing and maintaining compute‐and‐storage infrastructure to meet the demands of genomics as part of standard care. Policy on the storage of genomic data — either in a national genomic database or in state‐based systems — needs to be developed and is being informed by the work of Australian Genomics.
Curating sequence data
Variants identified by the bioinformatics pipeline through comparison with the reference genome need to be curated in the context of the patient's symptoms. The American College of Medical Genetics and Genomics and the Association for Molecular Pathology have published guidelines for interpreting sequence variants to determine the pathogenicity of particular variants.7 Scaling this curation step as genomics enters mainstream health care is complicated by current manual processes and specialised training of laboratory personnel. To meet the demands of scaling, the genetics workforce will need to be expanded, but big data analytics and computer‐based decision support will also be required. Sharing information on variants — including the underpinning evidence — is increasingly becoming an international priority.
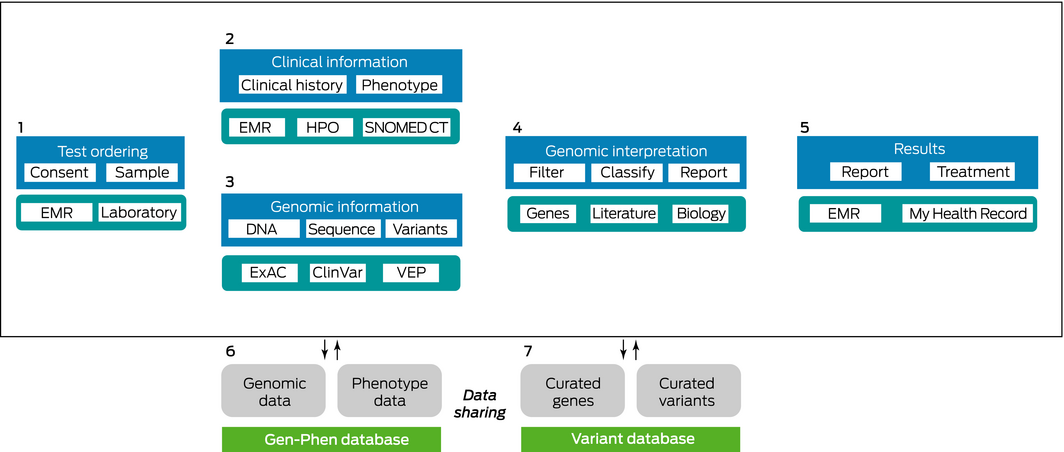
Genomic testing data flow
The flow of genomic data can be broken down into seven phases, shown in the Box. Phase 1 involves acquiring patient samples, with patient consent, in accredited clinical laboratories. This leads to phases 2 and 3, which happen in parallel. Phase 2 involves capturing relevant clinical information in electronic medical records using standardised terminology such as SNOMED CT and HPO, and phase 3 involves generating genomic information annotated with public information from sources such as ClinVar or the Exome Aggregation Consortium using tools such as Variant Effect Predictor. Phase 4 is the manual review and curation of annotated genomic changes relevant to the patient's phenotype. Phase 5 is generation of clinical reports for return to the electronic medical record and sharing via My Health Record. Phases 6 and 7 involve capturing generated results, annotation and relationships in dedicated databases for re‐use.
Genomics and digital health
Australia's national digital health strategy outlines the infrastructure, processes and policies that the federal government is putting in place to support Australia's transition to a digitally enabled health care system.8 Several interoperable pieces of infrastructure will be critical for supporting implementation of genomics into clinical care: a unique health care identifier for each patient and health care professional; a national clinical terminology; a shared health care record; and consistent national policies and legislation around data privacy, accessibility and linkage.
Combining health records and genomic information will mean that phenotypic data, as well as data on disease incidence, age at disease onset and life expectancy, can be used to inform clinical management. At a population level, the combined data will be useful for medical research as well as to enrich policy development and implementation, and economic modelling.
Ethics and consent
Genomics and digital health aim to ensure patients receive adequate information and opportunity to autonomously evaluate the risks, benefits and outcomes of health choices (including genomic testing) and thereby make informed choices. Genomic sequencing brings additional challenges of individual and family ownership of information.
To address these challenges, Australian Genomics is developing a national clinical genomic consent form that will introduce uniformity in the consent process for genomic testing and in materials used to educate patients and the community about genomics. In addition, several clinical projects are using a participant platform that incorporates a dynamic, granular consent model to inform and gauge patient decision making.9 This degree of autonomy and control to view and provide access to data is supported by the My Health Record system, which enables patients to share or restrict access to their data. These consent mechanisms will be important in providing patients with the ability to share their data anonymously while supplying the large scale data required to maximise the value of genomic medicine.
Use of genomics throughout life
Most clinical genetic or genomic activity to date has been managed in the secondary and tertiary health care sectors, being the remit of specialist and often multidisciplinary clinics. However, some genetic diseases (such as hereditary haemochromatosis, fragile X syndrome and familial hypercholesterolaemia) can be diagnosed and managed by general practitioners with the aid of a single gene test or chromosomal microarray.10 In the future, these single gene tests may be replaced by a laboratory test that interrogates the genomic variants in a patient's stored genomic sequence rather than requiring re‐sampling and repeat sequencing. Pharmacogenomics — the use of genomic data to identify the drug that will be safest and most effective at the individual patient level — will become especially important considering the burden of adverse drug events and ineffective drug use on the Australian health system.11
Conclusion
As the use of genomics increases, it will require additional infrastructure, processes and policies to ensure that it is accepted in the community and that it delivers on its promise of improved health outcomes. This is the remit of the National Health Genomics Policy Framework.12 Australian Genomics is working closely with a number of international genomic initiatives to ensure that knowledge and processes developed in one country can be transferred and utilised internationally. The symbiotic application and intersection with Australia's national digital health strategy will be crucial for supporting effective, ethical and sustainable integration of genomics into the Australian health care system.
The integration of knowledge about our genome can drive a personalised approach to whole‐of‐life care,13 but will necessarily require good electronic health records. For the optimal introduction of genomics into Australia's health care system, we need a fully functioning digital health system — supporting both patients and clinicians.
Provenance: Commissioned; externally peer reviewed.
- 1. Stark Z, Schofield D, Alam K, et al. Prospective comparison of the cost‐effectiveness of clinical whole‐exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med 2017; 19: 867–874.
- 2. Gaff CL, Winship IM, Forrest SM, et al. Preparing for genomic medicine: a real world demonstration of health system change. NPJ Genom Med 2017; 2: 16.
- 3. Global Alliance for Genomics and Health. Genomics. A federated ecosystem for sharing genomic, clinical data. Science 2016; 352: 1278–1280.
- 4. Donnelly K. SNOMED‐CT: the advanced terminology and coding system for ehealth. Stud Health Technol Inform 2006; 121: 279–290.
- 5. Robinson PN, Köhler S, Bauer S, et al. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet 2008; 83: 610–615.
- 6. Genome Reference Consortium [website]. https://www.ncbi.nlm.nih.gov/grc (viewed Apr 2018).
- 7. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424.
- 8. Australian Digital Health Agency. Australia's national digital health strategy. https://www.digitalhealth.gov.au/about-the-agency/publications/australias-national-digital-health-strategy (viewed Apr 2018).
- 9. Kaye J, Whitley EA, Lund D, et al. Dynamic consent: a patient interface for twenty‐first century research networks. Eur J Hum Genet 2015; 23: 141–146.
- 10. Royal Australian College of General Practitioners. Genomics in general practice. Melbourne: RACGP, 2018. https://www.racgp.org.au/your-practice/guidelines/genomics (viewed Apr 2018).
- 11. Miller GC, Britt HC, Valenti L. Adverse drug events in general practice patients in Australia. Med J Aust 2006; 184: 321–324. https://www.mja.com.au/journal/2006/184/7/adverse-drug-events-general-practice-patients-australia
- 12. Department of Health. National Health Genomics Policy Framework 2018–2021. http://www.health.gov.au/internet/main/publishing.nsf/Content/national-health-genomics-policy-framework-2018-2021 (viewed Apr 2018).
- 13. Bauer DC, Gaff C, Dinger ME, et al. Genomics and personalised whole‐of‐life healthcare. Trends Mol Med 2014; 20: 479–486.





The Australian Genomics Health Alliance is supported by the National Health and Medical Research Council (GNT1113531).
David Hansen is a member of the Clinical and Technical Board Advisory Committee of the Australian Digital Health Agency, and is employed by the CSIRO, which has partnered with the Australian Digital Health Agency to provide the National Clinical Terminology Service.